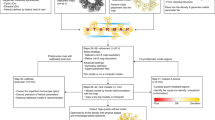
Abstract
For highly oriented fibrillar molecules, three-dimensional structures can often be determined from X-ray fiber diffraction data. However, because of limited information content, structure determination and validation can be challenging. We demonstrate that automated structure determination of protein fibers can be achieved by guiding the building of macromolecular models with fiber diffraction data. We illustrate the power of our approach by determining the structures of six bacteriophage viruses de novo using fiber diffraction data alone and together with solid-state NMR data. Furthermore, we demonstrate the feasibility of molecular replacement from monomeric and fibrillar templates by solving the structure of a plant virus using homology modeling and protein-protein docking. The generated models explain the experimental data to the same degree as deposited reference structures but with improved structural quality. We also developed a cross-validation method for model selection. The results highlight the power of fiber diffraction data as structural constraints.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chandrasekaran, R. & Stubbs., G. in International Tables for Crystallography Vol. F, 2nd edn. (eds. Arnold, E., Himmel, D.M. & Rossmann, M.G.) Ch. 19.5, 583–592 (International Union of Crystallography, 2012).
Brünger, A.T. Free R value: cross-validation in crystallography. Methods Enzymol. 277, 366–396 (1997).
Wang, H. & Stubbs, G. Structure determination of cucumber green mottle mosaic virus by X-ray fiber diffraction. Significance for the evolution of tobamoviruses. J. Mol. Biol. 239, 371–384 (1994).
Kaufmann, K.W., Lemmon, G.H., Deluca, S.L., Sheehan, J.H. & Meiler, J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry 49, 2987–2998 (2010).
Bradley, P., Misura, K.M.S. & Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871 (2005).
Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 (2003).
Welsh, L.C., Symmons, M.F. & Marvin, D.A. The molecular structure and structural transition of the á-helical capsid in filamentous bacteriophage Pf1. Acta Crystallogr. D Biol. Crystallogr. 56, 137–150 (2000).
Welsh, L.C., Symmons, M.F., Sturtevant, J.M., Marvin, D.A. & Perham, R.N. Structure of the capsid of Pf3 filamentous phage determined from X-ray fibre diffraction data at 3.1 Å resolution. J. Mol. Biol. 283, 155–177 (1998).
Wu, Y. & Ma, J. Refinement of F-actin model against fiber diffraction data by long-range normal modes. Biophys. J. 86, 116–124 (2004).
Makowski, L. The use of continuous diffraction data as a phase constraint. II. Application to fiber diffraction data. J. Appl. Crystallogr. 15, 546–557 (1982).
Tyka, M.D. et al. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 405, 607–618 (2011).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Falkner, B. & Schröder, G.F. Cross-validation in cryo-EM-based structural modeling. Proc. Natl. Acad. Sci. USA 110, 8930–8935 (2013).
Fabiola, F., Korostelev, A. & Chapman, M.S. Bias in cross-validated free R factors: mitigation of the effects of non-crystallographic symmetry. Acta Crystallogr. D Biol. Crystallogr. 62, 227–238 (2006).
Marvin, D.A., Welsh, L.C., Symmons, M.F., Scott, W.R.P. & Straus, S.K. Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J. Mol. Biol. 355, 294–309 (2006).
Straus, S.K., Scott, W.R.P., Schwieters, C.D. & Marvin, D.A. Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR. Eur. Biophys. J. 40, 221–234 (2011).
Straus, S.K., Scott, W.R.P., Symmons, M.F. & Marvin, D.A. On the structures of filamentous bacteriophage Ff (fd, f1, M13). Eur. Biophys. J. 37, 521–527 (2008).
Gonzalez, A., Nave, C. & Marvin, D.A. Pf1 filamentous bacteriophage: refinement of a molecular model by simulated annealing using 3.3 Å resolution X-ray fibre diffraction data. Acta Crystallogr. D Biol. Crystallogr. 51, 792–804 (1995).
Das, R. et al. Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl. Acad. Sci. USA 106, 18978–18983 (2009).
Zeri, A.C., Mesleh, M.F., Nevzorov, A.A. & Opella, S.J. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 100, 6458–6463 (2003).
Tewary, S.K. et al. Structure of hibiscus latent Singapore virus by fiber diffraction: a nonconserved His122 contributes to coat protein stability. J. Mol. Biol. 406, 516–526 (2011).
André, I., Bradley, P., Wang, C. & Baker, D. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. USA 104, 17656–17661 (2007).
Conway, P., Tyka, M.D., DiMaio, F., Konerding, D.E. & Baker, D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55 (2014).
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013).
Habeck, M., Rieping, W. & Nilges, M. Weighting of experimental evidence in macromolecular structure determination. Proc. Natl. Acad. Sci. USA 103, 1756–1761 (2006).
Beese, L., Stubbs, G. & Cohen, C. Microtubule structure at 18 Å resolution. J. Mol. Biol. 194, 257–264 (1987).
Nambudripad, R., Stark, W. & Makowski, L. Neutron diffraction studies of the structure of filamentous bacteriophage Pf1. J. Mol. Biol. 220, 359–379 (1991).
Brown, P.J., Fox, A.G., Maslen, E.N., O'Keefe, M.A. & Willis, B.T.M. in International Tables for Crystallography Vol. C (ed. Prince, E.) Ch. 6.1, 554–595 (International Union of Crystallography, 2006).
Keiner, J., Kunis, S. & Potts, D. Using NFFT 3—a software library for various nonequispaced fast Fourier transforms. ACM Trans. Math. Softw. 36, 19 (2009).
Siegman, A.E. Quasi fast Hankel transform. Opt. Lett. 1, 13 (1977).
DiMaio, F., Tyka, M.D., Baker, M.L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392, 181–190 (2009).
Abe, H., Braun, W., Noguti, T. & Gō´, N. Rapid calculation of first and second derivatives of conformational energy with respect to dihedral angles for proteins general recurrent equations. Comput. Chem. 8, 239–247 (1984).
Wedemeyer, W.J. & Baker, D. Efficient minimization of angle-dependent potentials for polypeptides in internal coordinates. Proteins 53, 262–272 (2003).
Rambo, R.P. & Tainer, J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
DiMaio, F., Leaver-Fay, A., Bradley, P., Baker, D. & André, I. Modeling symmetric macromolecular structures in Rosetta3. PLoS ONE 6, e20450 (2011).
Gront, D., Kulp, D.W., Vernon, R.M., Strauss, C.E.M. & Baker, D. Generalized fragment picking in Rosetta: design, protocols and applications. PLoS ONE 6, e23294 (2011).
Acknowledgements
This work was supported by the Crafoord Foundation and the Sven and Lilly Lawski Foundation. The simulations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at Lunarc. We thank S.K. Tewary (University of Kansas) for sharing fiber diffraction data of HLSV and input files for X-PLOR simulations. We thank S. Bjelic, S. Rämisch and R. Lizatovic for critically reading the manuscript.
Author information
Authors and Affiliations
Contributions
W.P. and I.A. designed the research, developed the methods and analyzed the data. W.P. carried out the simulations. W.P. and I.A. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Comparison between observed and calculated intensities.
For non-crystalline systems experimental fiber diffraction data (black) is confined to layer lines. Each layer is spaced along meridional direction at Z=l/c, where l is the layer line number and c is helical repeat; and sampled along the equator at an interval of R=0.0025 1/Å. Intensities were calculated from atomic coordinates in reciprocal space (blue, equation 5).
Supplementary Figure 2 Determining structures of bacteriophage viruses de novo with and without experimental data.
We ran Fold-And-Dock simulations guided by fiber diffraction data (blue) and in the absence of experimental data (green). The comparison is shown for: (a) Pf3 filamentous bacteriophage (PDB code: 1ifp), (b) filamentous bacteriophage PH75 (PDB code: 1hgv), (c) filamentous bacteriophage Pf1 (PDB code: 1ql1), (d) filamentous bacteriophage Pf1 (PDB code: 4ifm) and (e) fd filamentous bacteriophage (PDB code: 2c0w). (f) Additionally, fd filamentous bacteriophage (PDB code: 2c0x) simulations were guided by multiple experimental data sets (fiber diffraction and solid state NMR). Models generated without fiber diffraction were validated against work set with the Rosetta score application.
Supplementary Figure 3 Coarse-grained symmetrical docking of HLSV.
Coarse-grained symmetrical docking of (a) the monomeric structure of hibiscus latent Singapore virus (PDB code: 3pdm) and (b) the homology model based on the monomer of tobacco mosaic virus (PDB code: 2tmv). Blue dots corresponds to models obtained from docking guided by fiber diffraction data, while green for models generated without experimental restraints.
Supplementary Figure 4 CPU and GPU performance on calculations of all-atom intensity and derivatives.
All-atom intensity (equation 5) and derivatives (equations S5-S8) can be calculated sequentially using standard CPU or in parallel mode on a Graphical Processor Unit (GPU). We use a structure of hibiscus latent Singapore virus (HLSV) to compare the performance on both architectures. Monomer of HLSV has 162 amino-acid residues and was truncated by factor of 20 to perform calculations on models that consisted of 82, 102, 122 and 142 amino acid residues.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Table 1, Supplementary Notes 1 and 2 and Supplementary Protocol (PDF 1655 kb)
Supplementary Data 1
Protein models (ZIP 11329 kb)
Supplementary Data 2
Command files for running fiber diffraction modeling with Rosetta (ZIP 28624 kb)
Rights and permissions
About this article
Cite this article
Potrzebowski, W., André, I. Automated determination of fibrillar structures by simultaneous model building and fiber diffraction refinement. Nat Methods 12, 679–684 (2015). https://doi.org/10.1038/nmeth.3399
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3399