Abstract
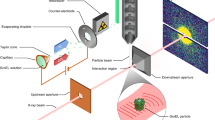
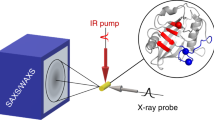
We describe a method for performing time-resolved X-ray crystallographic experiments based on the Hadamard transform, in which time resolution is defined by the underlying periodicity of the probe pulse sequence, and signal/noise is greatly improved over that for the fastest pump-probe experiments depending on a single pulse. This approach should be applicable on standard synchrotron beamlines and will enable high-resolution measurements of protein and small-molecule structural dynamics. It is also applicable to other time-resolved measurements where a probe can be encoded, such as pump-probe spectroscopy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
08 January 2015
In the version of this article initially published, the Figure 2 legend misidentified the control data as on the left and the HATRX data as on the right. The error has been corrected in the HTML and PDF versions of the article.
References
Copeland, R.A. Future Med. Chem. 3, 1491–1501 (2011).
Giuseppone, N., Fuks, G. & Lehn, J.-M. Chemistry 12, 1723–1735 (2006).
Buryak, A. & Severin, K. Angew. Chem. Int. Edn. Engl. 44, 7935–7938 (2005).
Zewail, A.H. in Femtochemistry: Ultrafast Dynamics of the Chemical Bond I (World Scientific, 1994).
Bourgeois, D. & Weik, M. Crystallogr. Rev. 15, 87–118 (2009).
Neutze, R. & Moffat, K. Curr. Opin. Struct. Biol. 22, 651–659 (2012).
Jung, Y.O. et al. Nat. Chem. 5, 212–220 (2013).
Schotte, F. et al. Proc. Natl. Acad. Sci. USA 109, 19256–19261 (2012).
Lindenberg, A.M. et al. Phys. Rev. Lett. 84, 111–114 (2000).
Beaud, P. et al. Phys. Rev. Lett. 103, 155702–155706 (2009).
Chapman, H.N. et al. Nature 470, 73–77 (2011).
Holton, J.M. & Frankel, K.A. Acta Crystallogr. D Biol. Crystallogr. 66, 393–408 (2010).
Moffat, K. Chem. Rev. 101, 1569–1581 (2001).
Hamm, P., Lim, M. & Hochstrasser, R.M. J. Chem. Phys. 102, 6123–6138 (1998).
Porter, G. Proc. R. Soc. Lond. A Math. Phys. Sci. 200, 284–300 (1950).
Harwit, M. & Sloane, N.J.A. in Hadamard Transform Optics. (Academic Press, 1979).
Fernández, F. et al. Anal. Chem. 74, 1611–1617 (2002).
Bolinger, L. & Leigh, J. J. Magn. Reson. 80, 162–167 (1988).
Garman, E.F. Acta Crystallogr. D Biol. Crystallogr. 66, 339–351 (2010).
Owen, R.L., Rudino-Pinera, E. & Garman, E.F. Proc. Natl. Acad. Sci. USA 103, 4912–4917 (2006).
Paithankar, K.S. & Garman, E.F. Acta Crystallogr. D Biol. Crystallogr. 66, 381–388 (2010).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011).
Waterman, D. & Evans, G. J. Appl. Cryst. 43, 1356–1371 (2010).
French, G.S. & Wilson, K.S. Acta Crystallogr. A 34, 517–525 (1978).
Sauter, C., Lorber, B. & Giegé, R. Proteins 48, 146–150 (2002).
Murshudov, G.N. et al. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Broennimann, C. et al. J. Synchrotron Radiat. 13, 120–130 (2006).
Cammarata, M. et al. Rev. Sci. Instrum. 80, 15101–15110 (2009).
Graber, T. et al. Synchrotron Radiat. 18, 658–670 (2011).
Nozawa, S. et al. J. Synchrotron Radiat. 14, 313–319 (2007).
Cole, J.M. Chem. Soc. Rev. 33, 501–513 (2004).
Sun, C., Portmann, G., Hertlein, M., Kirz, J. & Robin, D.S. Phys. Rev. Lett. 109, 264801–264805 (2012).
Acknowledgements
B.A.Y. is supported by the Wellcome Trust 4-year PhD program “The Molecular Basis of Biological Mechanisms” 089312/Z/09/Z. This work was also supported by the EPSRC Award “Dynamic Structural Science at the Research Complex at Harwell” EP/I01974X/1 and by BBSRC Award BB/H001905/1. Diffraction data were collected on beamline I24 at Diamond Light Source under proposal nt5810. We thank Diamond Light Source and the staff of I24 for beamtime and support and D. Waterman (CCP4) for the SUMSUB code. We also thank P. Raithby, C. Wilson, J. Trincao, J. Christensen, M. Hamilton, E. Snell, T. Burnley, C. Wood and C. Ballard for useful discussions and critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
G.S.B. and B.A.Y. originally proposed the application of the Hadamard transform to time-resolved experiments. B.A.Y., R.L.O. and A.R.P. devised the proof-of-principle crystallographic experiment and collected the data. B.A.Y., R.L.O., G.S.B. and A.R.P. all processed data, wrote software and jointly wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 4 and 5. (PDF 10730 kb)
Supplementary Table 1
Scaling statistics for the reference data (XLSX 102 kb)
Supplementary Table 2
Scaling statistics for the n = 3 HATRX data (XLSX 93 kb)
Supplementary Table 3
Weighted R factors (W rel) by resolution shell14 for the comparison of the reference data (R) at each time point with the HATRX data. (XLSX 56 kb)
Supplementary Software 1
This script takes a user defined set of images from multiple crystals and produces an integrated list of reflections using XDS. These data can then be scaled, merged and converted to structure factors using the CCP4 software suite. (TXT 10 kb)
Supplementary Software 2
This script takes a plain text file (with the format H K L F1 SIGF1 F2 SIGF2 ... Fn SIGFn), calculates the S-matrix and carries out the HATRX transform. It outputs a plain text file in the same format as the input data. (TXT 3 kb)
Supplementary Software 3
This script converts the output of 2_HATRX.py into a CCP4 format mtz file ready for further analysis using any standard crystallographic software package. (TXT 0 kb)
Supplementary Software 4
This script uses the sumsub routine to create HATRX encoded images from a series of CBF format images. It was used to create the HATRX images here to show proof of principle of the method. When the HATRX data are generated by detector or beam encoding this step is not required. (TXT 3 kb)
Rights and permissions
About this article
Cite this article
Yorke, B., Beddard, G., Owen, R. et al. Time-resolved crystallography using the Hadamard transform. Nat Methods 11, 1131–1134 (2014). https://doi.org/10.1038/nmeth.3139
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3139
This article is cited by
-
LED-pump-X-ray-multiprobe crystallography for sub-second timescales
Communications Chemistry (2022)
-
A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density
Nature Communications (2017)
-
Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons
Nature Communications (2017)
-
Protein structure through time
Nature Methods (2016)
-
Erratum: Corrigendum: Time-resolved crystallography using the Hadamard transform
Nature Methods (2015)