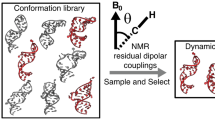
Abstract
We present a simple and general approach termed REsemble for quantifying population overlap and structural similarity between ensembles. This approach captures improvements in the quality of ensembles determined using increasing input experimental data—improvements that go undetected when conventional methods for comparing ensembles are used—and reveals unexpected similarities between RNA ensembles determined using NMR and molecular dynamics simulations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jensen, M.R. et al. Structure 17, 1169–1185 (2009).
Clore, G.M. & Schwieters, C.D. Biochemistry 43, 10678–10691 (2004).
Salmon, L., Yang, S. & Al-Hashimi, H.M. Annu. Rev. Phys. Chem. 10.1146/annurev-physchem-040412-110059 (16 December 2013).
Boehr, D.D., Nussinov, R. & Wright, P.E. Nat. Chem. Biol. 5, 789–796 (2009).
Wand, A.J. Curr. Opin. Struct. Biol. 23, 75–81 (2013).
Stelzer, A.C. et al. Nat. Chem. Biol. 7, 553–559 (2011).
Richardson, J.S. & Richardson, D.C. Annu. Rev. Biophys. 42, 1–28 (2013).
Lindorff-Larsen, K. & Ferkinghoff-Borg, J. PLoS ONE 4, e4203 (2009).
Fisher, C.K., Huang, A. & Stultz, C.M. J. Am. Chem. Soc. 132, 14919–14927 (2010).
De Simone, A., Richter, B., Salvatella, X. & Vendruscolo, M. J. Am. Chem. Soc. 131, 3810–3811 (2009).
Cha, S.-H. Int. J. Math. Models Methods Appl. Sci. 1, 300–307 (2007).
Brüschweiler, R. Curr. Opin. Struct. Biol. 13, 175–183 (2003).
Frank, A.T., Stelzer, A.C., Al-Hashimi, H.M. & Andricioaei, I. Nucleic Acids Res. 37, 3670–3679 (2009).
Marsh, J.A., Teichmann, S.A. & Forman-Kay, J.D. Curr. Opin. Struct. Biol. 22, 643–650 (2012).
Chen, Y., Campbell, S.L. & Dokholyan, N.V. Biophys. J. 93, 2300–2306 (2007).
Tolman, J.R., Flanagan, J.M., Kennedy, M.A. & Prestegard, J.H. Proc. Natl. Acad. Sci. USA 92, 9279–9283 (1995).
Tjandra, N. & Bax, A. Science 278, 1111–1114 (1997).
Bailor, M.H., Mustoe, A.M., Brooks, C.L. III. & Al-Hashimi, H.M. Nat. Protoc. 6, 1536–1545 (2011).
Salmon, L., Bascom, G., Andricioaei, I. & Al-Hashimi, H.M. J. Am. Chem. Soc. 135, 5457–5466 (2013).
Denning, E.J., Priyakumar, U.D., Nilsson, L. & Mackerell, A.D. Jr. J. Comput. Chem. 32, 1929–1943 (2011).
Fisher, C.K., Zhang, Q., Stelzer, A. & Al-Hashimi, H.M. J. Phys. Chem. B 112, 16815–16822 (2008).
Lavery, R. & Sklenar, H. J. Biomol. Struct. Dyn. 6, 655–667 (1989).
Acknowledgements
We thank members of the Al-Hashimi laboratory for critical comments on the manuscript. This work was supported by the US National Institutes of Health (R01AI066975 and PO1 GM0066275).
Author information
Authors and Affiliations
Contributions
S.Y., L.S. and H.M.A.-H. conceived the idea; S.Y. and L.S. carried out the data analysis with help from H.M.A.-H.; S.Y., L.S. and H.M.A.-H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
H.M.A.-H. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1631 kb)
Supplementary Software
Source code of SAS and ensemble similarity measurement (ZIP 95 kb)
Source data
Rights and permissions
About this article
Cite this article
Yang, S., Salmon, L. & Al-Hashimi, H. Measuring similarity between dynamic ensembles of biomolecules. Nat Methods 11, 552–554 (2014). https://doi.org/10.1038/nmeth.2921
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2921
This article is cited by
-
DNA mismatches reveal conformational penalties in protein–DNA recognition
Nature (2020)
-
High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble
Nature Structural & Molecular Biology (2018)
-
Conformational ensemble comparison for small molecules in drug discovery
Journal of Computer-Aided Molecular Design (2018)
-
Integrative, dynamic structural biology at atomic resolution—it's about time
Nature Methods (2015)