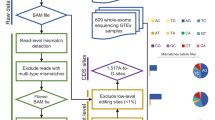
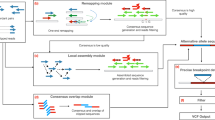
Abstract
We show that RNA editing sites can be called with high confidence using RNA sequencing data from multiple samples across either individuals or species, without the need for matched genomic DNA sequence. We identified many previously unidentified editing sites in both humans and Drosophila; our results nearly double the known number of human protein recoding events. We also found that human genes harboring conserved editing sites within Alu repeats are enriched for neuronal functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Nishikura, K. Annu. Rev. Biochem. 79, 321–349 (2010).
Kim, U., Wang, Y., Sanford, T., Zeng, Y. & Nishikura, K. Proc. Natl. Acad. Sci. USA 91, 11457–11461 (1994).
Levanon, E.Y. et al. Nat. Biotechnol. 22, 1001–1005 (2004).
Eisenberg, E., Li, J.B. & Levanon, E.Y. RNA Biol. 7, 248–252 (2010).
Rosenthal, J.J. & Seeburg, P.H. Neuron 74, 432–439 (2012).
Bahn, J.H. et al. Genome Res. 22, 142–150 (2012).
Peng, Z. et al. Nat. Biotechnol. 30, 253–260 (2012).
Ramaswami, G. et al. Nat. Methods 9, 579–581 (2012).
DePristo, M.A. et al. Nat. Genet. 43, 491–498 (2011).
Li, M. et al. Science 333, 53–58 (2011).
Kleinman, C.L., Adoue, V. & Majewski, J. RNA 18, 1586–1596 (2012).
Kleinman, C.L. & Majewski, J. Science 335, 1302 (2012).
Lin, W., Piskol, R., Tan, M.H. & Li, J.B. Science 335, 1302 (2012).
Pickrell, J.K., Gilad, Y. & Pritchard, J.K. Science 335, 1302 (2012).
Schrider, D.R., Gout, J.F. & Hahn, M.W. PLoS ONE 6, e25842 (2011).
Piskol, R., Peng, Z., Wang, J. & Li, J.B. Nat. Biotechnol. 31, 19–20 (2013).
Li, J.B. et al. Science 324, 1210–1213 (2009).
Goodman, M. Am. J. Hum. Genet. 64, 31–39 (1999).
Paz-Yaacov, N. et al. Proc. Natl. Acad. Sci. USA 107, 12174–12179 (2010).
Tamura, K., Subramanian, S. & Kumar, S. Mol. Biol. Evol. 21, 36–44 (2004).
Hoopengardner, B., Bhalla, T., Staber, C. & Reenan, R. Science 301, 832–836 (2003).
Huang da, W. Nat. Protoc. 4, 44–57 (2009).
Li, H. & Durbin, R. Bioinformatics 26, 589–595 (2010).
Vacic, V., Iakoucheva, L.M. & Radivojac, P. Bioinformatics 22, 1536–1537 (2006).
Kiran, A. & Baranov, P.V. Bioinformatics 26, 1772–1776 (2010).
Palladino, M.J., Keegan, L.P., O'Connell, M.A. & Reenan, R.A. Cell 102, 437–449 (2000).
Picardi, E. et al. Nucleic Acids Res. 38, 4755–4767 (2010).
Acknowledgements
We thank E. Levanon, A. Fire and members of the Li lab for constructive discussions, S. Blair for assistance with data set curation, and modENCODE Consortium for the use of Drosophila RNA-seq data sets. G.R. and P.D. were supported by the Stanford Genome Training Program funded by the US National Institutes of Health. R.Z. was partially supported by a Dean's fellowship from Stanford University School of Medicine. R.P. was supported by a fellowship from the German Academic Exchange Service. This work was supported by startup funds from Stanford University Department of Genetics and Ellison Medical Foundation (to J.B.L.) and Medical Research Council, UK (to M.A.O.).
Author information
Authors and Affiliations
Contributions
G.R. and R.Z. performed computational analyses with help from R.P., P.D. and J.B.L.; R.Z. and G.R. carried out the validation experiments; L.P.K. and M.A.O. generated RNA-seq data for wild-type and Adar5G1 flies; and G.R., R.Z. and J.B.L. wrote the paper with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–19, Supplementary Tables 1–10, Supplementary Notes 1–5 (PDF 2289 kb)
Supplementary Data 1
A-to-G mismatches identified in lymphoblastoid cell lines (XLSX 23087 kb)
Supplementary Data 2
Non-A-to-G mismatches identified in lymphoblastoid cell lines (XLSX 8502 kb)
Supplementary Data 3
A-to-G mismatches identified in human brain tissues (XLSX 50144 kb)
Supplementary Data 4
Non-A-to-G mismatches identified in human brain tissues (XLSX 11477 kb)
Supplementary Data 5
A-to-G mismatches identified in other human tissues (XLSX 31524 kb)
Supplementary Data 6
Non-A-to-G mismatches identified in other human tissues (XLSX 9841 kb)
Supplementary Data 7
A-to-G mismatches identified in primate cross-species comparison (XLSX 1479 kb)
Supplementary Data 8
Non-A-to-G mismatches identified in primate cross-species comparison (XLSX 160 kb)
Supplementary Data 9
A-to-G mismatches identified in Drosophila cross-species comparison (XLSX 70 kb)
Supplementary Data 10
Non-A-to-G mismatches identified in Drosophila cross-species comparison (XLSX 35 kb)
Rights and permissions
About this article
Cite this article
Ramaswami, G., Zhang, R., Piskol, R. et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 10, 128–132 (2013). https://doi.org/10.1038/nmeth.2330
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2330
This article is cited by
-
Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus
BMC Biology (2023)
-
L-GIREMI uncovers RNA editing sites in long-read RNA-seq
Genome Biology (2023)
-
A novel computational method enables RNA editome profiling during human hematopoiesis from scRNA-seq data
Scientific Reports (2023)
-
Evidence Supporting That C-to-U RNA Editing Is the Major Force That Drives SARS-CoV-2 Evolution
Journal of Molecular Evolution (2023)
-
The role of post-transcriptional modifications during development
Biologia Futura (2023)