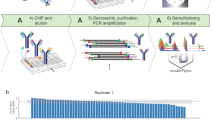
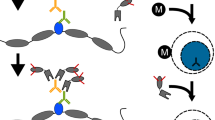
Abstract
Genome-wide profiling of transcription factors based on massive parallel sequencing of immunoprecipitated chromatin (ChIP-seq) requires nanogram amounts of DNA. Here we describe a high-fidelity, single-tube linear DNA amplification method (LinDA) for ChIP-seq and reChIP-seq with picogram DNA amounts obtained from a few thousand cells. This amplification technology will facilitate global analyses of transcription-factor binding and chromatin with very small cell populations, such as stem or cancer-initiating cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Acevedo, L.G. et al. Biotechniques 43, 791–797 (2007).
O'Neill, L.P., VerMilyea, M.D. & Turner, B.M. Nat. Genet. 38, 835–841 (2006).
Dahl, J.A. & Collas, P. Nat. Protoc. 3, 1032–1045 (2008).
O'Geen, H., Nicolet, C.M., Blahnik, K., Green, R. & Farnham, P.J. Biotechniques 41, 577–580 (2006).
Pfeifer, G.P., Steigerwald, S.D., Mueller, P.R., Wold, B. & Riggs, A.D. Science 246, 810–813 (1989).
Pinard, R. et al. BMC Genomics 7, 216 (2006).
Talseth-Palmer, B.A., Bowden, N.A., Hill, A., Meldrum, C. & Scott, R.J. BMC Res. Notes 1, 56 (2008).
Goren, A. et al. Nat. Methods 7, 47–49 (2010).
Liu, C.L., Schreiber, S.L. & Bernstein, B.E. BMC Genomics 4, 19 (2003).
Adli, M., Zhu, J. & Bernstein, B.E. Nat. Methods 7, 615–618 (2010).
van Bakel, H. et al. Nucleic Acids Res. 36, e21 (2008).
Liu, C.L., Bernstein, B.E. & Schreiber, S.L. Cold Spring Harb. Protoc. pdb.top42 (2008).
Ceschin, D.G. et al. Genes Dev. (in the press).
Zhang, Y. et al. Genome Biol. 9, R137 (2008).
Ye, T. et al. Nucleic Acids Res. 39, e35 (2011).
Acknowledgements
We thank B. Jost, S. Vicaire and S. Le Gras for Illumina sequencing and mapping to the genomes, W. van Gool for help in the data analysis, A. Krebs for help with seqMINER and D. Ceschin advice on statistics and for critical reading of the manuscript. P.S. is a postdoc at the Ligue Nationale Contre le Cancer; M.W. is supported by fellowships of the Association pour le Recherche sur le Cancer and the Fondation pour la Recherche Médicale. This work was supported by funds from the Ligue Nationale Contre le Cancer (laboratoire labelisé) and European Community contracts LSHC-CT-2005-518417 'EPITRON', LSHG-CT2005-018882 'X-TRA-NET', LSHM-CT2005-018652 'CRESCENDO' and HEALTH-F4-2009-221952 'ATLAS'.
Author information
Authors and Affiliations
Contributions
P.S., L.M.T. and H.G. designed and optimized LinDA. M.-A.M.-P. performed the F9-cell experiments and did the bioinformatics analysis with P.S., and M.W. did the H3396-cell experiments. N.L. and L.W. performed the Illumina HiSeq 2000 sequencing and mapping to hg19 human genome assembly. H.G. and P.S. wrote the main text of the manuscript, which M.-A.M.-P. and L.M.T. corrected and improved; P.S. and L.M.T. wrote the Online Methods section.
Corresponding author
Ethics declarations
Competing interests
H.G., P.S. and L.M.T. filed a patent application describing the methods presented here (European patent EP11305531.3).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–2 (PDF 880 kb)
Rights and permissions
About this article
Cite this article
Shankaranarayanan, P., Mendoza-Parra, MA., Walia, M. et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods 8, 565–567 (2011). https://doi.org/10.1038/nmeth.1626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1626
This article is cited by
-
High-throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers
BMC Genomics (2019)
-
FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation
BMC Biology (2019)
-
Terminal transfer amplification and sequencing for high-efficiency and low-bias copy number profiling of fragmented DNA samples
Protein & Cell (2019)
-
Cell Type-Specific Survey of Epigenetic Modifications by Tandem Chromatin Immunoprecipitation Sequencing
Scientific Reports (2018)
-
Purification of nanogram-range immunoprecipitated DNA in ChIP-seq application
BMC Genomics (2017)