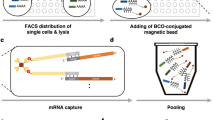
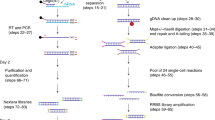
Abstract
Generating reliable expression profiles from minute cell quantities is critical for scientific discovery and potential clinical applications. Here we present low-quantity digital gene expression (LQ-DGE), an amplification-free approach involving capture of poly(A)+ RNAs from cellular lysates onto poly(dT)-coated sequencing surfaces, followed by on-surface reverse transcription and sequencing. We applied LQ-DGE to profile malignant and nonmalignant mouse and human cells, demonstrating its quantitative power and potential applicability to archival specimens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Linsen, S.E. et al. Nat. Methods 6, 474–476 (2009).
Pinard, R. et al. BMC Genomics 7, 216 (2006).
Subkhankulova, T. & Livesey, F.J. Genome Biol. 7, R18 (2006).
Taniguchi, K., Kajiyama, T. & Kambara, H. Nat. Methods 6, 503–506 (2009).
Lipson, D. et al. Nat. Biotechnol. 27, 652–658 (2009).
Pushkarev, D., Neff, N.F. & Quake, S.R. Nat. Biotechnol. 27, 847–852 (2009).
Aguirre, A.J. et al. Genes Dev. 17, 3112–3126 (2003).
Bardeesy, N. et al. Proc. Natl. Acad. Sci. USA 103, 5947–5952 (2006).
Ozsolak, F. et al. Nature 461, 814–818 (2009).
Slomovic, S., Laufer, D., Geiger, D. & Schuster, G. Nucleic Acids Res. 34, 2966–2975 (2006).
Perocchi, F., Xu, Z., Clauder-Munster, S. & Steinmetz, L.M. Nucleic Acids Res. 35, e128 (2007).
Ozsolak, F. et al. Genome Res. 20, 519–525 (2010).
Oshlack, A. & Wakefield, M.J. Biol. Direct 4, 14 (2009).
Mamanova, L. et al. Nat. Methods 7, 130–132 (2010).
Dennis, G. et al. Genome Biol. 4, 3 (2003).
Cleveland, W.S. J. Am. Stat. Assoc. 74, 829–836 (1979).
Acknowledgements
We thank K. Kerouac for technical assistance. D.T.T. is supported by the Pancreatic Cancer Action Network–American Association for Cancer Research Research Fellowship, and D.A.H. is supported by the Howard Hughes Medical Institute. This work was supported by US National Human Genome Research Institute grant 1R44HG005279-01 to F.O. and P.M.M.
Author information
Authors and Affiliations
Contributions
F.O. conceived and designed the study; F.O., D.T.T., B.W.B. and S.P. performed the experiments; N.B. provided the cells; F.O., B.S.W. and S.R. performed the computational analyses; F.O., D.T.T., P.M.M. and D.A.H. wrote the paper, which all authors edited.
Corresponding authors
Ethics declarations
Competing interests
F.O. and P.M.M. are employees of Helicos BioSciences Corporation.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1–4 and Supplementary Discussion (PDF 2211 kb)
Rights and permissions
About this article
Cite this article
Ozsolak, F., Ting, D., Wittner, B. et al. Amplification-free digital gene expression profiling from minute cell quantities. Nat Methods 7, 619–621 (2010). https://doi.org/10.1038/nmeth.1480
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1480