Abstract
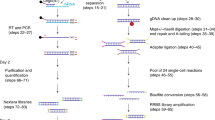
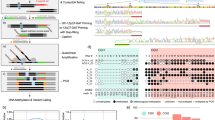
Bisulfite sequencing measures absolute levels of DNA methylation at single-nucleotide resolution, providing a robust platform for molecular diagnostics. We optimized bisulfite sequencing for genome-scale analysis of clinical samples: here we outline how restriction digestion targets bisulfite sequencing to hotspots of epigenetic regulation and describe a statistical method for assessing significance of altered DNA methylation patterns. Thirty nanograms of DNA was sufficient for genome-scale analysis and our protocol worked well on formalin-fixed, paraffin-embedded samples.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Esteller, M. Nat. Rev. Genet. 8, 286–298 (2007).
Lister, R. & Ecker, J.R. Genome Res. 19, 959–966 (2009).
Meissner, A. et al. Nucleic Acids Res. 33, 5868–5877 (2005).
Meissner, A. et al. Nature 454, 766–770 (2008).
Down, T.A. et al. Nat. Biotechnol. 26, 779–785 (2008).
Serre, D., Lee, B.H. & Ting, A.H. Nucleic Acids Res. published online, doi:10.1093/nar/gkp992 (11 November 2009).
Brunner, A.L. et al. Genome Res. 19, 1044–1056 (2009).
Irizarry, R.A. et al. Genome Res. 18, 780–790 (2008).
Irizarry, R.A. et al. Nat. Genet. 41, 178–186 (2009).
Lister, R. et al. Cell 133, 523–536 (2008).
Bock, C., Halachev, K., Büch, J. & Lengauer, T. Genome Biol. 10, R14 (2009).
Bibikova, M. et al. Epigenomics 1, 177 (2009).
Bock, C. et al. Nucleic Acids Res. 36, e55 (2008).
Eckhardt, F. et al. Nat. Genet. 38, 1378–1385 (2006).
Hellebrekers, D.M. et al. Clin. Cancer Res. 15, 3990–3997 (2009).
Zhang, W. et al. Cancer Res. 68, 2764–2772 (2008).
Weisenberger, D.J. et al. Nat. Genet. 38, 787–793 (2006).
Garrison, W.D. et al. Gastroenterology 130, 1207–1220 (2006).
Bock, C. Epigenomics 1, 99 (2009).
Smith, Z.D., Gu, H., Bock, C., Gnirke, A. & Meissner, A. Methods 48, 226 (2009).
Storey, J.D. & Tibshirani, R. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Acknowledgements
We thank K. Halachev (Max Planck Institute for Informatics) for providing genome annotation files and H. Cedar (The Hebrew University of Jerusalem) for providing the human blood DNA samples. C.B. is supported by a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation. A.M. is supported by the Massachusetts Life Science Center and the Pew Charitable Trusts. The described work was in part funded by US National Institutes of Health grants R01HG004401, U54HG03067 and U01ES017155.
Author information
Authors and Affiliations
Contributions
H.G., C.B., A.G., E.S.L. and A.M. conceived and designed the experiments; H.G. and E.T. performed the experiments; C.B. analyzed data; T.S.M., N.J. and Z.D.S. contributed materials or analysis tools; and H.G., C.B. and A.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2 and Supplementary Table 1 (PDF 822 kb)
Supplementary Note
Source code of Epigenome pipeline package with documentation and demonstration data. (ZIP 88748 kb)
Rights and permissions
About this article
Cite this article
Gu, H., Bock, C., Mikkelsen, T. et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 7, 133–136 (2010). https://doi.org/10.1038/nmeth.1414
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1414
This article is cited by
-
Broodstock nutritional programming differentially affects the hepatic transcriptome and genome-wide DNA methylome of farmed gilthead sea bream (Sparus aurata) depending on genetic background
BMC Genomics (2023)
-
Identification and validation of a DNA methylation-driven gene-based prognostic model for clear cell renal cell carcinoma
BMC Genomics (2023)
-
DNA methylation profiling to determine the primary sites of metastatic cancers using formalin-fixed paraffin-embedded tissues
Nature Communications (2023)
-
DNA methylation and expression profiles of placenta and umbilical cord blood reveal the characteristics of gestational diabetes mellitus patients and offspring
Clinical Epigenetics (2022)
-
Genome-wide DNA methylation patterns reveal clinically relevant predictive and prognostic subtypes in human osteosarcoma
Communications Biology (2022)