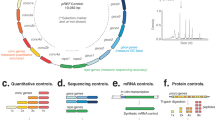
Abstract
Appropriate resources and expression technology necessary for human proteomics on a whole-proteome scale are being developed. We prepared a foundation for simple and efficient production of human proteins using the versatile Gateway vector system. We generated 33,275 human Gateway entry clones for protein synthesis, developed mRNA expression protocols for them and improved the wheat germ cell-free protein synthesis system. We applied this protein expression system to the in vitro expression of 13,364 human proteins and assessed their biological activity in two functional categories. Of the 75 tested phosphatases, 58 (77%) showed biological activity. Several cytokines containing disulfide bonds were produced in an active form in a nonreducing wheat germ cell-free expression system. We also manufactured protein microarrays by direct printing of unpurified in vitro–synthesized proteins and demonstrated their utility. Our 'human protein factory' infrastructure includes the resources and expression technology for in vitro proteome research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Ota, T. et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45 (2004).
Kimura, K. et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16, 55–65 (2006).
Otsuki, T. et al. Signal sequence and keyword trap in silico for selection of full-length human cDNAs encoding secretion or membrane proteins from oligo-capped cDNA libraries. DNA Res. 12, 117–126 (2005).
Strausberg, R.L. et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 99, 16899–16903 (2002).
Wiemann, S. et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 11, 422–435 (2001).
Nomura, N. et al. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1, 251–262 (1994).
Temple, G. et al. From genome to proteome: developing expression clone resources for the human genome. Hum. Mol. Genet. 15, R31–43 (2006).
Hartley, J.L., Temple, G.F. & Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 (2000).
Sawasaki, T., Ogasawara, T., Morishita, R. & Endo, Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 99, 14652–14657 (2002).
Endo, Y. & Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 17, 373–380 (2006).
Sawasaki, T. et al. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 514, 102–105 (2002).
Hirokawa, T., Boon-Chieng, S. & Mitaku, S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 (1998).
Takano, E. et al. Pig heart calpastatin: identification of repetitive domain structures and anomalous behavior in polyacrylamide gel electrophoresis. Biochemistry 27, 1964–1972 (1988).
Robinson, N.C. & Tanford, C. The binding of deoxycholate, Triton X-100, sodium dodecyl sulfate, and phosphatidylcholine vesicles to cytochrome b5. Biochemistry 14, 369–378 (1975).
Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004).
Moorhead, G.B., Trinkle-Mulcahy, L. & Ulke-Lemee, A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8, 234–244 (2007).
Taylor, G.S., Maehama, T. & Dixon, J.E. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 97, 8910–8915 (2000).
Wishart, M.J. & Dixon, J.E. PTEN and myotubularin phosphatases: from 3-phosphoinositide dephosphorylation to disease. Trends Cell Biol. 12, 579–585 (2002).
Honda, R., Ohba, Y., Nagata, A., Okayama, H. & Yasuda, H. Dephosphorylation of human p34cdc2 kinase on both Thr-14 and Tyr-15 by human cdc25B phosphatase. FEBS Lett. 318, 331–334 (1993).
Nguyen, T.H., Liu, J. & Lombroso, P.J. Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J. Biol. Chem. 277, 24274–24279 (2002).
Somani, A.K. et al. The SH2 domain containing tyrosine phosphatase-1 down-regulates activation of Lyn and Lyn-induced tyrosine phosphorylation of the CD19 receptor in B cells. J. Biol. Chem. 276, 1938–1944 (2001).
Kawasaki, T., Gouda, M.D., Sawasaki, T., Takai, K. & Endo, Y. Efficient synthesis of a disulfide-containing protein through a batch cell-free system from wheat germ. Eur. J. Biochem. 270, 4780–4786 (2003).
Miura, A. et al. Differential responses of normal human coronary artery endothelial cells against multiple cytokines comparatively assessed by gene expression profiles. FEBS Lett. 580, 6871–6879 (2006).
Ito, E. et al. A tetraspanin-family protein, T-cell acute lymphoblastic leukemia-associated antigen 1, is induced by the Ewing's sarcoma-Wilms' tumor 1 fusion protein of desmoplastic small round-cell tumor. Am. J. Pathol. 163, 2165–2172 (2003).
Sakamoto, A. et al. Influence of inhalation anesthesia assessed by comprehensive gene expression profiling. Gene 356, 39–48 (2005).
Jobling, S.A. et al. Biological activity and receptor binding of human prointerleukin-1 beta and subpeptides. J. Biol. Chem. 263, 16372–16378 (1988).
Miranda-Saavedra, D. & Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins 68, 893–914 (2007).
Lowell, C.A. Src-family kinases: rheostats of immune cell signaling. Mol. Immunol. 41, 631–643 (2004).
Lamesch, P. et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307–315 (2007).
Zhu, H. et al. Global analysis of protein activities using proteome chips. Science 293, 2101–2105 (2001).
Plutzky, J., Neel, B.G. & Rosenberg, R.D. Isolation of a src homology 2–containing tyrosine phosphatase. Proc. Natl. Acad. Sci. USA 89, 1123–1127 (1992).
Acknowledgements
This work was supported by the grant 'Functional Analysis of Human Proteins and Its Research Application' from the New Energy and Industrial Technology Development Organization (NEDO), Japan. We thank the Research Association for Biotechnology and Helix Research Institute for providing FLJ cDNA clones, S. Sugano (Graduate School of Frontier Sciences, The University of Tokyo) for providing the oligo-capped cDNA libraries, helpful suggestions and encouragement during the work, A. Miyawaki (Brain Science Institute, RIKEN) for providing the plasmid encoding the Venus protein, and S. Ebisu (ProteinExpress, Co., Ltd.), H. Takakura (Takara) and M. Hirano (Toray Research Center, Inc.) for providing E. coli cells for protein expression systems.
Author information
Authors and Affiliations
Contributions
N.G., Yo.K, Ya.K., S.T., Ka.M., A.W., T.I., S.W. and N.N. designed and supervised the project. Ay.M., Yo.K., J.-i.I., N.G. and N.N. wrote the manuscript. T.I., A.W., J.-i.Y., Ko.K., T.N., T.A. and R.K. determined ORF regions and selected templates. T.A., N.K., R.K., K.I, At.M., T.O., Ki.K., Yuk.S.,Ts.S., S.S., Ya.K., K.Y., N.S. and S.T. produced Gateway entry clones. Y.I., K.-i.K., Y.T. and Ki.M. sequenced Gateway entry clones. K.Y., R.S., Y.M., C.K. and N.S. re-examined identity and stability of Gateway entry clones at the final step. B.K, A.S., T.K., Mar.M., H.T. and N.K. produced Gateway destination vectors. R.M., Ay.M., A.F., Yut.S., To.S., Mas.M., To.T., H.K. and H.T. performed preliminary analysis for determining best conditions for protein production and other experimental protocols. A.F., Yut.S., H.Y., B.K., A.S., T.K., S.N. and M.K. expressed proteins and performed SDS-PAGE. A.F., To.S., To.T., Mas.M. and Yo.K. analyzed the SDS-PAGE data. A.F., Ay.M. and Yo.K. analyzed phosphatase activity. Ay.M., Ta.T., R.H., Y.Y., E.I., A.N. and J.-i.I. analyzed cytokine activity by DNA microarrays. A.F., H.Y., C.K., Yut.S, Yo.K. and J.-i.I. produced protein microarrays. A.F., Yo.K. and J.-i.I. analyzed data of protein microarrays. F.I. and Y.E. made comments and encouraged the idea.
Corresponding authors
Ethics declarations
Competing interests
Ko.K. and T.N. are employees of Hitachi. Y.I., K.-i.K. and K.Y. are employees of Hitachi Science Systems. B.K. and T.K. are employees of Toyobo. M.K. is an employee of Mitsubishi Chemical Group. S.N. is an employee of Wakenyaku.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1, 3–7 and 9, Supplementary Methods, Supplementary Note (PDF 3498 kb)
Supplementary Table 2
Features of all Gateway standard entry and its source cDNA clones. (TXT 2162 kb)
Supplementary Table 8
List of 13,277 Gateway entry clones whose proteins were mounted on protein microarrays and fluorescence intensity of each spot. (TXT 865 kb)
Rights and permissions
About this article
Cite this article
Goshima, N., Kawamura, Y., Fukumoto, A. et al. Human protein factory for converting the transcriptome into an in vitro–expressed proteome. Nat Methods 5, 1011–1017 (2008). https://doi.org/10.1038/nmeth.1273
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1273
This article is cited by
-
Identification of G protein-coupled receptor 55 (GPR55) as a target of curcumin
npj Science of Food (2022)
-
CF-PPiD technology based on cell-free protein array and proximity biotinylation enzyme for in vitro direct interactome analysis
Scientific Reports (2022)
-
A novel family of expression vectors with multiple affinity tags for wheat germ cell-free protein expression
BMC Biotechnology (2020)
-
Reconstitution of Drosophila and human chromatins by wheat germ cell-free co-expression system
BMC Biotechnology (2020)
-
UHRF1-KAT7-mediated regulation of TUSC3 expression via histone methylation/acetylation is critical for the proliferation of colon cancer cells
Oncogene (2020)