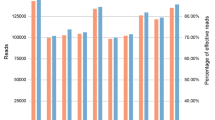
Abstract
We developed a generalized framework for multiplexed resequencing of targeted human genome regions on the Illumina Genome Analyzer using degenerate indexed DNA bar codes ligated to fragmented DNA before sequencing. Using this method, we simultaneously sequenced the DNA of multiple HapMap individuals at several Encyclopedia of DNA Elements (ENCODE) regions. We then evaluated the use of Bayes factors for discovering and genotyping polymorphisms. For polymorphisms that were either previously identified within the Single Nucleotide Polymorphism database (dbSNP) or visually evident upon re-inspection of archived ENCODE traces, we observed a false positive rate of 11.3% using strict thresholds for predicting variants and 69.6% for lax thresholds. Conversely, false negative rates were 10.8–90.8%, with false negatives at stricter cut-offs occurring at lower coverage (<10 aligned reads). These results suggest that >90% of genetic variants are discoverable using multiplexed sequencing provided sufficient coverage at the polymorphic base.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Zondervan, K.T. & Cardon, L.R. Designing candidate gene and genome-wide case-control association studies. Nat. Protoc. 2, 2492–2501 (2007).
Meyer, M., Stenzel, U., Myles, S., Prüfer, K. & Hofreiter, M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 35, e97 (2007).
Parameswaran, P. et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35, e130 (2007).
Milosavljevic, A. et al. Pooled genomic indexing of rhesus macaque. Genome Res. 15, 292–301 (2005).
Hamady, M., Walker, J.J., Harris, J.K., Gold, N.J. & Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5, 235–237 (2008).
ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Albert, T.J. et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods 4, 903–905 (2007).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527 (2007).
Porreca, G.J. et al. Multiplex amplification of large sets of human exons. Nat. Methods 4, 931–936 (2007).
Okou, D.T. et al. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods 4, 907–909 (2007).
Jeck, W.R. et al. Extending assembly of short DNA sequences to handle error. Bioinformatics 23, 2942–2944 (2007).
Acknowledgements
We acknowledge funding from the state of Arizona, US National Heart Lung and Blood Institute (U01 HL086528), the Stardust foundation, Science Foundation Arizona, and National Institute for Neurological Disorders and Strokes (R01 N5059873).
Author information
Authors and Affiliations
Contributions
D.W.C., J.V.P., M.J.H., G.N. and D.A.S. contributed to initial experimental design. S.S., A.S., M.R., J.J.C., T.L. and T.L.P. contributed to development and execution of exact experimental protocols. J.V.P., D.W.C. and N.H. contributed to the development of bioinformatics and analysis pipelines.
Corresponding author
Ethics declarations
Competing interests
G.N. is an employee of Illumina.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Tables 1–5, Supplementary Methods (PDF 434 kb)
Rights and permissions
About this article
Cite this article
Craig, D., Pearson, J., Szelinger, S. et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods 5, 887–893 (2008). https://doi.org/10.1038/nmeth.1251
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1251
This article is cited by
-
Population admixtures in medaka inferred by multiple arbitrary amplicon sequencing
Scientific Reports (2022)
-
Analysis of allelic variants of RhMLO genes in rose and functional studies on susceptibility to powdery mildew related to clade V homologs
Theoretical and Applied Genetics (2021)
-
Skim sequencing: an advanced NGS technology for crop improvement
Journal of Genetics (2021)
-
Sequencing barcode construction and identification methods based on block error-correction codes
Science China Life Sciences (2020)
-
Effects of Heat stress and molecular mitigation approaches in orphan legume, Chickpea
Molecular Biology Reports (2020)