Abstract
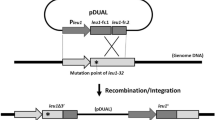
Here we describe a facile plasmid-chromosome shuffling technique for generating and analyzing non-deletion alleles in the yeast Saccharomyces cerevisiae. This technique takes advantage of an existing set of genome-wide haploid-convertible heterozygous diploid yeast knockout mutants. This simple method will facilitate characterization of essential gene functions and genome-wide investigation of protein structure-function relationships.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Boeke, J.D., Trueheart, J., Natsoulis, G. & Fink, G.R. Methods Enzymol. 154, 164–175 (1987).
Budd, M. & Campbell, J.L. Proc. Natl. Acad. Sci. USA 84, 2838–2842 (1987).
Muhlrad, D., Hunter, R. & Parker, R. Yeast 8, 79–82 (1992).
Pan, X. et al. Cell 124, 1069–1081 (2006).
Pan, X. et al. Mol. Cell 16, 487–496 (2004).
Tong, A.H. et al. Science 294, 2364–2368 (2001).
Giaever, G. et al. Nature 418, 387–391 (2002).
Amin, N.S. & Holm, C. Genetics 144, 479–493 (1996).
Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G. & Jentsch, S. Nature 419, 135–141 (2002).
Lau, P.J., Flores-Rozas, H. & Kolodner, R.D. Mol. Cell. Biol. 22, 6669–6680 (2002).
Krishna, T.S., Kong, X.P., Gary, S., Burgers, P.M. & Kuriyan, J. Cell 79, 1233–1243 (1994).
Acknowledgements
We thank S. Kron (University of Chicago) for advice in implementing the 96-well yeast plasmid recovery protocol. This work was supported by a Baylor College of Medicine seed grant to X.P. and US National Institutes of Health (NIH) Roadmap grant “Technology Center for Networks and Pathways” (RR020839) to J.D.B. R.S.S. is supported by NIH P01 grant HD39691.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–6, Supplementary Methods (PDF 398 kb)
Rights and permissions
About this article
Cite this article
Huang, Z., Sucgang, R., Lin, Yy. et al. Plasmid-chromosome shuffling for non-deletion alleles in yeast. Nat Methods 5, 167–169 (2008). https://doi.org/10.1038/nmeth.1173
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1173