Abstract
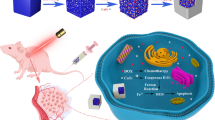
Conventional cancer therapies involve the systemic delivery of anticancer agents that neither discriminate between cancer and normal cells nor eliminate the risk of cancer recurrence. Here, we demonstrate that the combination of gene, drug and phototherapy delivered through a prophylactic hydrogel patch leads, in a colon cancer mouse model, to complete tumour remission when applied to non-resected tumours and to the absence of tumour recurrence when applied following tumour resection. The adhesive hydrogel patch enhanced the stability and provided local delivery of embedded nanoparticles. Spherical gold nanoparticles were used as a first wave of treatment to deliver siRNAs against Kras, a key oncogene driver, and rod-shaped gold nanoparticles mediated the conversion of near-infrared radiation into heat, causing the release of a chemotherapeutic as well as thermally induced cell damage. This local, triple-combination therapy can be adapted to other cancer cell types and to molecular targets associated with disease progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 (2016).
Kistler, C. E. Colorectal-cancer incidence and mortality after screening. N. Engl. J. Med. 369, 2354–2354 (2013).
Kountouras, J., Zavos, C. & Chatzopoulos, D. Therapy for colorectal cancer. N. Engl. J. Med. 352, 1820–1821 (2005).
Minelli, C., Lowe, S. B. & Stevens, M. M. Engineering nanocomposite materials for cancer therapy. Small 6, 2336–2357 (2010).
Liu, Y., Miyoshi, H. & Nakamura, M. Nanomedicine for drug delivery and imaging: a promising avenue for cancer therapy and diagnosis using targeted functional nanoparticles. Int. J. Cancer 120, 2527–2537 (2007).
Conde, J., Oliva, N., Atilano, M., Song, H. S. & Artzi, N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nature Mater. 15, 353–363 (2016).
Howes, P. D., Chandrawati, R. & Stevens, M. M. Colloidal nanoparticles as advanced biological sensors. Science 346, 1247390 (2014).
Wu, S. Y., Lopez-Berestein, G., Calin, G. A. & Sood, A. K. RNAi therapies: drugging the undruggable. Sci. Transl. Med. 6, 240ps7 (2014).
Bertrand, N., Wu, J., Xu, X., Kamaly, N. & Farokhzad, O. C. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 66, 2–25 (2014).
Conde, J., Edelman, E. R. & Artzi, N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: the jack-of-all-trades in cancer nanotheranostics? Adv. Drug Deliv. Rev. 81, 169–183 (2015).
Oliva, N. et al. Regulation of dendrimer/dextran material performance by altered tissue microenvironment in inflammation and neoplasia. Sci. Transl. Med. 7, 272ra211 (2015).
Segovia, N. et al. Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Adv. Healthc. Mater. 4, 271–280 (2015).
Conde, J., Oliva, N. & Artzi, N. Implantable hydrogel embedded dark-gold nanoswitch as a theranostic probe to sense and overcome cancer multidrug resistance. Proc. Natl Acad. Sci. USA 112, E1278–E1287 (2015).
Artzi, N. et al. In vivo and in vitro tracking of erosion in biodegradable materials using non-invasive fluorescence imaging. Nature Mater. 10, 704–709 (2011).
Artzi, N., Shazly, T., Baker, A. B., Bon, A. & Edelman, E. R. Aldehyde-amine chemistry enables modulated biosealants with tissue-specific adhesion. Adv. Mater. 21, 3399–3403 (2009).
von Maltzahn, G. et al. Computationally guided photothermal tumor therapy using long-circulating gold nanorod antennas. Cancer Res. 69, 3892–3900 (2009).
Huang, X., El-Sayed, I. H., Qian, W. & El-Sayed, M. A. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J. Am. Chem. Soc. 128, 2115–2120 (2006).
von, M. G. et al. Nanoparticles that communicate in vivo to amplify tumour targeting. Nature Mater. 10, 545–552 (2011).
Sonpavde, G. Bevacizumab in colorectal cancer. N. Engl. J. Med. 351, 1690 (2004).
Wang, Y., Fei, D., Vanderlaan, M. & Song, A. Biological activity of bevacizumab, a humanized anti-VEGF antibody in vitro. Angiogenesis 7, 335–345 (2004).
Li, Z. J. et al. A novel peptide specifically targeting the vasculature of orthotopic colorectal cancer for imaging detection and drug delivery. J. Control. Release 148, 292–302 (2010).
Sunakawa, Y., Lenz, H. J. & Ichikawa, W. RAS mutations in colorectal cancer. N. Engl. J. Med. 369, 2159 (2013).
Bosch, F. X., Garten, W., Klenk, H. D. & Rott, R. Proteolytic cleavage of influenza-virus hemagglutinins—primary structure of the connecting peptide between Ha1 and Ha2 determines proteolytic cleavability and pathogenicity of avian influenza-viruses. Virology 113, 725–735 (1981).
Oliva, N. et al. Natural tissue microenvironmental conditions modulate adhesive material performance. Langmuir 28, 15402–15409 (2012).
Wang, Y. H. et al. Synergistic delivery of gold nanorods using multifunctional microbubbles for enhanced plasmonic photothermal therapy. Sci. Rep. 4, 5685 (2014).
Kuo, W. S. et al. Gold nanorods in photodynamic therapy, as hyperthermia agents, and in near-infrared optical imaging. Angew. Chem. Int. Ed. Engl. 49, 2711–2715 (2010).
Chong, P. S. & Finlay, I. G. Surgical options in the management of advanced and recurrent colorectal cancer. Surg. Oncol. 16, 25–31 (2007).
Chokshi, R. J., Abdel-Misih, S. & Bloomston, M. Surgical management of colorectal cancer: a review of the literature. Indian J. Surg. 71, 350–355 (2009).
Mocellin, S. Microarray Technology and Cancer Gene Profiling. Advances in Experimental Medicine and Biology (Springer, 2007).
Adiconis, X. et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nature Methods 10, 623–629 (2013).
Hemmat, M. et al. Short stature, digit anomalies and dysmorphic facial features are associated with the duplication of miR-17 ∼ 92 cluster. Mol. Cytogenet. 7, 27 (2014).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Mendell, J. T. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222 (2008).
Hatakeyama, H. et al. A DNA microarray-based analysis of the host response to a nonviral gene carrier: a strategy for improving the immune response. Mol. Ther. 19, 1487–1498 (2011).
Meerzaman, D. M. et al. Genome-wide transcriptional sequencing identifies novel mutations in metabolic genes in human hepatocellular carcinoma. Cancer Genomics Proteomics 11, 1–12 (2014).
Voloshanenko, O. et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nature Commun. 4, 2610 (2013).
Herath, N. I., Doecke, J., Spanevello, M. D., Leggett, B. A. & Boyd, A. W. Epigenetic silencing of EphA1 expression in colorectal cancer is correlated with poor survival. Br. J. Cancer 100, 1095–1102 (2009).
Kok-Sin, T. et al. Identification of diagnostic markers in colorectal cancer via integrative epigenomics and genomics data. Oncol. Rep. 34, 22–32 (2015).
Sturn, A., Quackenbush, J. & Trajanoski, Z. Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208 (2002).
Mootha, V. K. et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
J.C. acknowledges Marie Curie International Outgoing Fellowship and Funding (FP7-PEOPLE-2013-IOF, Project 626386). We thank the Swanson Biotechnology Center at the Koch Institute for Integrative Cancer Research at Massachusetts Institute of Technology (MIT) for assistance with animal experiments and facilities, especially the microscopy, flow cytometry, and histology cores. We also acknowledge that all qPCR and microarray experiments done in the KI Genomics Core/MIT BioMicro Center are funded by the NIH and supported in part by the Koch Institute Support Grant P30-CA14051 from the National Cancer Institute and by the National Institute of Environmental Health Sciences of the NIH under award P30-ES002109. We thank D. Ma and C. Whittaker from the Bioinformatics and Computing Core Facility at David H. Koch Institute for Integrative Cancer Research at MIT for the microarrays analysis. We thank D. S. Yun for cryo-TEM assistance at the Peterson Nanotechnology Materials Core Facility. We thank the Department of Comparative Medicine at MIT, especially J. Haupt. We thank G. Paradis for FACS assistance with Cancer Center Support (FACS core) Grant P30-CA14051 from the National Cancer Institute. We thank P. Boisvert and Y. Zhang for technical assistance in SEM at the MIT Center for Materials Science and Engineering (CMSE). These SEM studies made use of the MRSEC Shared Experimental Facilities at MIT, supported by the National Science Foundation under award number DMR-1419807.
Author information
Authors and Affiliations
Contributions
J.C. and N.A. conceived the project and designed the experiments. J.C. and N.O. performed the experiments, and collected and analysed the data. Y.Z. performed the SEM studies. J.C. and N.A. co-wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary information (PDF 4166 kb)
Rights and permissions
About this article
Cite this article
Conde, J., Oliva, N., Zhang, Y. et al. Local triple-combination therapy results in tumour regression and prevents recurrence in a colon cancer model. Nature Mater 15, 1128–1138 (2016). https://doi.org/10.1038/nmat4707
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat4707
This article is cited by
-
Atomically precise photothermal nanomachines
Nature Materials (2024)
-
Hydrogels for RNA delivery
Nature Materials (2023)
-
Multi-bioinspired Sprayable Nanotherapeutics for Tumor-Specific Focal Cancer Therapy
Biotechnology and Bioprocess Engineering (2023)
-
New anti-tumor strategy based on acid-triggered self-destructive and near-infrared laser light responses of nano-biocatalysts integrating starvation–chemo–photothermal therapies
Cancer Nanotechnology (2022)
-
Injectable magnetic montmorillonite colloidal gel for the postoperative treatment of hepatocellular carcinoma
Journal of Nanobiotechnology (2022)