Abstract
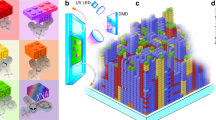
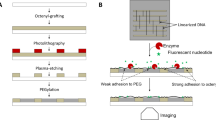
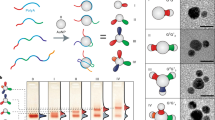
As demonstrated by means of DNA nanoconstructs1, as well as DNA functionalization of nanoparticles2,3,4 and micrometre-scale colloids5,6,7,8, complex self-assembly processes require components to associate with particular partners in a programmable fashion. In many cases the reversibility of the interactions between complementary DNA sequences is an advantage9. However, permanently bonding some or all of the complementary pairs may allow for flexibility in design and construction10. Here, we show that the substitution of a cinnamate group for a pair of complementary bases provides an efficient, addressable, ultraviolet light-based method to bond complementary DNA covalently. To show the potential of this approach, we wrote micrometre-scale patterns on a surface using ultraviolet light and demonstrated the reversible attachment of conjugated DNA and DNA-coated colloids. Our strategy enables both functional DNA photolithography and multistep, specific binding in self-assembly processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996).
Nykypanchuk, D., Maye, M. M., van der Lelie, D & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Biancaniello, P. L., Kim, A. J. & Crocker, J. C. Colloidal interactions and self-assembly using DNA hybridization. Phys. Rev. Lett. 94, 058302 (2005).
Valignat, M. P., Theodoly, O., Crocker, J. C., Russel, W. B. & Chaikin, P. M. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc. Natl Acad. Sci. USA 102, 4225–4229 (2005).
Rogers, P. H. et al. Controllable, and reversible aggregation of polystyrene latex microspheres via DNA hybridization. Langmuir 21, 5562–5569 (2005).
Leunissen, M. E. et al. Switchable self-protected attractions in DNA-functionalized colloids. Nature Mater. 8, 590–595 (2009).
Wang, T. et al. Self-replication of information-bearing nanoscale patterns. Nature 478, 225–228 (2011).
Leunissen, M. E. et al. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter 5, 2422–2430 (2009).
Kallenbach, N. R., Ma, R-I. & Seeman, N. C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 305, 829–831 (1983).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Macfarlane, R. J. et al. Nanoparticle superlattice engineering with DNA. Science 334, 204–208 (2011).
Winfree, E., Liu, F. R., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Sherman, W. B. & Seeman, N. C. DNA walking biped. Nano Lett. 4, 1203–1207 (2004).
Yurke, B., Turberfield, A. J., Mills, A. P. J., Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Lewin, D. I. DNA computing. Comput. Sci. Eng. 4, 5–8 (2002).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Schulman, R. & Winfree, E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc. Natl Acad. Sci. USA 104, 15236–15241 (2007).
Gu, H., Chao, J., Xiao, S. J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010).
Wassarman, D. & Steitz, J. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257, 1918–1925 (1992).
Takasugi, M. et al. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc. Natl Acad. Sci. USA 88, 5602–5606 (1991).
Wu, Q., Christensen, L. A., Legerski, R. J. & Vasquez, K. M. Mismatch repair participates in error-free processing of DNA interstrand cross-links in human cells. EMBO Rep. 6, 551–557 (2005).
Yoshimura, Y, Itoa, Y. & Fujimotoa, K. Interstrand photocross-linking of DNA via p-carbamoylvinyl phenol nucleoside. Bioorg. Med. Chem. Lett. 15, 1299–1301 (2005).
Yoshimura, Y. & Fujimoto, K. Ultrafast reversible photo-cross-linking reaction: Toward in situ DNA manipulation. Org. Lett. 10, 3227–3230 (2008).
Dreyfus, R. et al. Simple quantitative model for the reversible association of DNA coated colloids. Phys. Rev. Lett. 102, 048301 (2009).
Xu, Q., Feng, L., Sha, R., Seeman, N. C. & Chaikin, P. M. Subdiffusion of a sticky particle on a surface. Phys. Rev. Lett. 106, 228102 (2011).
Xia, D., Yan, J. & Hou, S. Fabrication of nanofluidic biochips with nanochannels for applications in DNA analysis. Small 8, 2787–2801 (2012).
Maalouf, A., Gadonna, M. & Bosc, D. An improvement in standard photolithography resolution based on Kirchhoff diffraction studies. J. Phys. D 42, 015106 (2009).
Gorzolnik, B., Mela, P. & Möller, M. Nano-structured micropatterns by combination of block copolymer self-assembly and UV photolithography. Nanotechnology 17, 5027–5032 (2006).
Varghese, B. et al. Size selective assembly of colloidal particles on a template by directed self-assembly technique. Langmuir 22, 8248–8252 (2006).
Naiser, T., Mai, T., Michel, W. & Ott, A. Versatile maskless microscope projection photolithography system and its application in light-directed fabrication of DNA microarrays. Rev. Sci. Instrum. 77, 063711 (2006).
Chee, M. et al. Accessing genetic information with high-density DNA arrays. Science 274, 610–614 (1996).
Liu, Q. H. et al. DNA computing on surfaces. Nature 403, 175–179 (2000).
Pregibon, D. C., Toner, M. & Doyle, P. S. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 315, 1393–1396 (2007).
Jiang, S. et al. Janus particle synthesis and assembly. Adv. Mater. 22, 1060–1071 (2010).
Caruthers, M. H. Gene synthesis machines: DNA chemistry and its uses. Science 230, 281–85 (1985).
Acknowledgements
This research has been partially supported by the MRSEC Program of the National Science Foundation under Award Number DMR-0820341 for the cinnamate-functionalized phosphoramidite, NASA NNX08AK04G for microscopy, and DOE-BES-DE-SC0007991 to P.C. for data acquisition and analysis, as well as by the following grants to N.C.S. for DNA synthesis and characterization: GM-29554 from the National Institute of General Medical Sciences, CTS-0608889 and CCF-0726378 from the National Science Foundation, 48681-EL and W911NF-07-1-0439 from the Army Research Office, and N000140910181 and N000140911118 from the Office of Naval Research. J. Romulus acknowledges support through the Margaret Strauss Kramer Graduate Student Fellowship in Chemistry.
Author information
Authors and Affiliations
Contributions
L.F. designed and performed experiments, analysed data and wrote the paper; J. Romulus and M.L. synthesized the cinnamate-containing phosphoramidite and wrote the paper; R.S. incorporated the cinnamate in the DNA strands, performed gel experiments, analysed data and wrote the paper; J. Royer performed experiments, analysed data and wrote the paper; K-T.W. performed experiments, analysed data and wrote the paper; Q.X. performed experiments and analysed data; N.C.S. initiated and directed the project, designed experiments, analysed data and wrote the paper; M.W. initiated and directed the project, designed experiments and wrote the paper; P.C. initiated and directed the project, designed experiments, analysed data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 780 kb)
Supplementary Information
Supplementary Movie S1 (AVI 2417 kb)
Supplementary Information
Supplementary Movie S2 (AVI 3713 kb)
Supplementary Information
Supplementary Movie S3 (AVI 6058 kb)
Rights and permissions
About this article
Cite this article
Feng, L., Romulus, J., Li, M. et al. Cinnamate-based DNA photolithography. Nature Mater 12, 747–753 (2013). https://doi.org/10.1038/nmat3645
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat3645
This article is cited by
-
Multi-level patterning nucleic acid photolithography
Nature Communications (2019)
-
DNA-Coated Microspheres and Their Colloidal Superstructures
Macromolecular Research (2018)
-
Three-dimensional super-resolution longitudinal magnetization spot arrays
Light: Science & Applications (2017)
-
Re-entrant solidification in polymer–colloid mixtures as a consequence of competing entropic and enthalpic attractions
Nature Materials (2015)
-
Reconfigurable multi-scale colloidal assembly on excluded volume patterns
Scientific Reports (2015)