Abstract
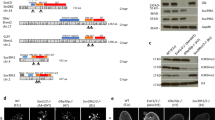
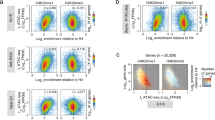
Studies of histone methylation have shown that H3 can be methylated at lysine 4 (Lys4) or lysine 9 (Lys9)1,2. Whereas H3–Lys4 methylation has been correlated with active gene expression3, H3–Lys9 methylation has been linked to gene silencing and assembly of heterochromatin in mouse and Schizosaccharomyces pombe4,5,6,7. The chromodomain of mouse HP1 (and Swi6 in S. pombe) binds H3 methylated at Lys9, and methylation at this site is thought to mark and promote heterochromatin assembly. We have used a well-studied model of mammalian epigenetic silencing, the human inactive X chromosome, to show that enrichment for H3 methylated at Lys9 is also a distinguishing mark of facultative heterochromatin. In contrast, H3 methylated at Lys4 is depleted in the inactive X chromosome, except in three 'hot spots' of enrichment along its length. Chromatin immunoprecipitation analyses further show that Lys9 methylation is associated with promoters of inactive genes, whereas Lys4 methylation is associated with active genes on the X chromosome. These data demonstrate that differential methylation at two distinct sites of the H3 amino terminus correlates with contrasting gene activities and may be part of a 'histone code' involved in establishing and maintaining facultative heterochromatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rice, J.C. & Allis, C.D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell. Biol. 13, 263–273 (2001).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Strahl, B.D., Ohba, R., Cook, R.G. & Allis, C.D. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in tetrahymena. Proc. Natl Acad. Sci. USA 96, 14967–14972 (1999).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bannister, A.J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Jacobs, S.A. et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232–5241 (2001).
Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D. & Grewal, S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Avner, P. & Heard, E. X-chromosome inactivation: counting, choice and initiation. Nature Rev. Genet. 2, 59–67 (2001).
Morishima, A., Grumbach, M. M. & Taylor, J.H. Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc. Natl Acad. Sci. USA 48, 756–763 (1962).
Barr, M.L. & Carr, D.H. Correlation between sex chromatin and chromosomes. Acta Cytol. 6, 34–45 (1962).
Norris, D.P., Brockdorff, N. & Rastan, S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm. Genome 1, 78–83 (1991).
Jeppesen, P. & Turner, B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74, 281–289 (1993).
Belyaev, N., Keohane, A.M. & Turner, B.M. Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum. Genet. 97, 573–578 (1996).
Boggs, B.A., Connors, B., Sobel, R.E., Chinault, A.C. & Allis, C.D. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma 105, 303–309 (1996).
Costanzi, C. & Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Chadwick, B.P. & Willard, H.F. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 10, 1101–1113 (2001).
Csankovszki, G., Nagy, A. & Jaenisch, R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153, 773–784 (2001).
Gartler, S.M. & Riggs, A.D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190 (1983).
Perche, P.Y. et al. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10, 1531–1534 (2000).
Francke, U. Digitized and differentially shaded human chromosome ideograms for genomic applications. Cytogenet. Cell Genet. 65, 206–218 (1994).
Carrel, L., Cottle, A.A., Goglin, K.C. & Willard, H.F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA 96, 14440–14444 (1999).
Ellis, N., Keitges, E., Gartler, S.M. & Rocchi, M. High-frequency reactivation of X-linked genes in Chinese hamster X human hybrid cells. Somat. Cell Mol. Genet. 13, 191–204 (1987).
Gilbert, S.L. & Sharp, P.A. Promoter-specific hypoacetylation of X-inactivated genes. Proc. Natl Acad. Sci. USA 96, 13825–13830 (1999).
Peters, A.H.F.M. et al. Histone H3 lysine 9 methylation is an epigenetic imprint for facultative heterochromatin. Nature Genet. DOI: 10.1038/ng 789 (2002).
Heard, E. et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X-inactivation. Cell (in the press).
Litt, M.D., Simpson, M., Gaszner, M., Allis, C.D. & Felsenfeld, G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293, 2453–2455 (2001).
Noma, K.I., Allis, C.D. & Grewal, S.I. Transitions in distinct histone h3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155 (2001).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Clemson, C.M., McNeil, J.A., Willard, H.F. & Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275 (1996).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Acknowledgements
We thank M.A. Jelinek and R. Rice (Upstate Biotechnology) for technical assistance in generating the methyl H3-specific antibodies, and A. Baldini for helpful discussions. This work was funded by National Institutes of Health grants to C.D.A., A.C.C. and D.L.S. E.H. was supported by NATO and the Centre National de la Recherche Scientifique.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boggs, B., Cheung, P., Heard, E. et al. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30, 73–76 (2002). https://doi.org/10.1038/ng787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng787
This article is cited by
-
BAF complex-mediated chromatin relaxation is required for establishment of X chromosome inactivation
Nature Communications (2022)
-
Laboratory methods to decipher epigenetic signatures: a comparative review
Cellular & Molecular Biology Letters (2021)
-
Deletion of the XIST promoter from the human inactive X chromosome compromises polycomb heterochromatin maintenance
Chromosoma (2021)
-
Characterization of chromatin at structurally abnormal inactive X chromosomes reveals potential evidence of a rare hybrid active and inactive isodicentric X chromosome
Chromosome Research (2020)
-
Varying levels of X chromosome coalescence in female somatic cells alters the balance of X-linked dosage compensation and is implicated in female-dominant systemic lupus erythematosus
Scientific Reports (2019)