Abstract
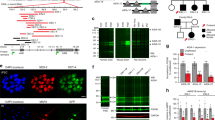
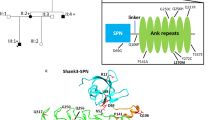
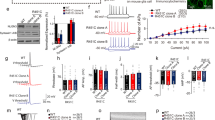
SHANK3 (also known as ProSAP2) regulates the structural organization of dendritic spines and is a binding partner of neuroligins; genes encoding neuroligins are mutated in autism and Asperger syndrome. Here, we report that a mutation of a single copy of SHANK3 on chromosome 22q13 can result in language and/or social communication disorders. These mutations concern only a small number of individuals, but they shed light on one gene dosage–sensitive synaptic pathway that is involved in autism spectrum disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Folstein, S.E. & Rosen-Sheidley, B. Nat. Rev. Genet. 2, 943–955 (2001).
Persico, A.M. & Bourgeron, T. Trends Neurosci. 29, 349–358 (2006).
Vorstman, J.A. et al. Mol. Psychiatry 11, 1, 18–28 (2006).
Manning, M.A. et al. Pediatrics 114, 451–457 (2004).
Bonaglia, M.C. et al. J. Med. Genet. 43, 822–828 (2006).
Naisbitt, S. et al. Neuron 23, 569–582 (1999).
Boeckers, T.M., Bockmann, J., Kreutz, M.R. & Gundelfinger, E.D. J. Neurochem. 81, 903–910 (2002).
Meyer, G., Varoqueaux, F., Neeb, A., Oschlies, M. & Brose, N. Neuropharmacology 47, 724–733 (2004).
Jamain, S. et al. Nat. Genet. 34, 27–29 (2003).
Bonaglia, M.C. et al. Am. J. Hum. Genet. 69, 261–268 (2001).
Roussignol, G. et al. J. Neurosci. 25, 3560–3570 (2005).
Baron, M.K. et al. Science 311, 531–535 (2006).
Lai, C.S., Fisher, S.E., Hurst, J.A., Vargha-Khadem, F. & Monaco, A.P. Nature 413, 519–523 (2001).
Somerville, M.J. et al. N. Engl. J. Med. 353, 1694–1701 (2005).
Carlisle, H.J. & Kennedy, M.B. Trends Neurosci. 28, 182–187 (2005).
Acknowledgements
We thank the affected individuals and their families for participating in this study and all the collaborators of the Paris Autism Research International Sibpair Study: C. Gillberg, M. Råstam, I.C. Gillberg, G. Nygren, H. Anckarsäter and O. Ståhlberg (Department of Child and Adolescent Psychiatry, Göteborg University); M. Leboyer (Department of Psychiatry, Groupe Hospitalier Albert Chenevier et Henri Mondor); C. Betancur (INSERM U513, Université Paris XII); C. Colineaux, D. Cohen, N. Chabane and M.-C. Mouren-Siméoni (Service de Psychopathologie de l'Enfant et l'Adolescent, Hôpital Robert Debré); A. Brice (INSERM U679, Hôpital Pitié-Salpêtrière); E. Sponheim (Centre for Child and Adolescent Psychiatry, University of Oslo); O.H. Skjeldal (Department of Pediatrics, Rikshospitalet, University of Oslo); M. Coleman (Department of Pediatrics, Georgetown University School of Medicine); P.L. Pearl (Children's National Medical Center, George Washington University School of Medicine); I.L. Cohen and J. Tsiouris (New York State Institute for Basic Research in Developmental Disabilities); Michele Zappella (Divisione di Neuropsichiatria Infantile, Azienda Ospedaliera Senese); H. Aschauer (Department of General Psychiatry, University Hospital, Vienna) and L. Van Maldergem (Centre de Génétique Humaine, Institut de Pathologie et de Génétique). We also thank the DNA and cell bank of INSERM U679 (IFR des Neurosciences, Hôpital Pitié-Salpêtrière); the Centre d'Investigations Cliniques of the Hôpital Robert Debré; C. Bouchier and S. Duthoy for the use of sequencing facilities at the Génopole Pasteur and A. Hchikat, L. Margarit and G. Rouffet for technical assistance. This work was supported by the Pasteur Institute, INSERM, Assistance Publique-Hôpitaux de Paris, Fondation France Télécom, Cure Autism Now, Fondation de France, Fondation Biomédicale de la Mairie de Paris, Fondation pour la Recherche Médicale, EUSynapse European Commission FP6, AUTISM MOLGEN European Commission FP6, Fondation NRJ, the Swedish Science Council and the Deutsche Forschungsgemeinschaft DFG, SFB 497.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Genomic structure and mRNA expression of the human SHANK3 gene. (PDF 1208 kb)
Supplementary Fig. 2
Genomic sequence of the deletion breakpoint in family ASD 1 and prediction of quadruplex-forming G-rich sequences (QGRS) at the terminal end of chromosome 22q13. (PDF 118 kb)
Supplementary Fig. 3
Pedigree structure, haplotype analyses and conservation of the SHANK3 mutations and variants identified in individuals with autism. (PDF 495 kb)
Supplementary Fig. 4
Analyses of SHANK3 mutations in rat hippocampal neuronal cultures. (PDF 168 kb)
Supplementary Table 1
SHANK3 variations identified in families with ASD and controls. (PDF 39 kb)
Supplementary Table 2
Primers used in this study. (PDF 43 kb)
Rights and permissions
About this article
Cite this article
Durand, C., Betancur, C., Boeckers, T. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 39, 25–27 (2007). https://doi.org/10.1038/ng1933
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1933
This article is cited by
-
Rare genetic brain disorders with overlapping neurological and psychiatric phenotypes
Nature Reviews Neurology (2024)
-
Combined expansion and STED microscopy reveals altered fingerprints of postsynaptic nanostructure across brain regions in ASD-related SHANK3-deficiency
Molecular Psychiatry (2024)
-
Deficiency of FRMD5 results in neurodevelopmental dysfunction and autistic-like behavior in mice
Molecular Psychiatry (2024)
-
The Shank/ProSAP N-Terminal (SPN) Domain of Shank3 Regulates Targeting to Postsynaptic Sites and Postsynaptic Signaling
Molecular Neurobiology (2024)
-
Description of a patient with developmental delay and dysmorphic features caused by a novel SHANK2 deletion
Egyptian Journal of Medical Human Genetics (2023)