Abstract
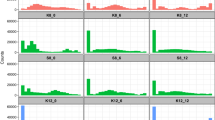
The budding yeast Saccharomyces cerevisiae has been used by humans for millennia to make wine, beer and bread1. More recently, it became a key model organism for studies of eukaryotic biology and for genomic analysis2. However, relatively little is known about the natural lifestyle and population genetics of yeast3. One major question is whether genetically diverse yeast strains mate and recombine in the wild. We developed a method to infer the evolutionary history of a species from genome sequences of multiple individuals and applied it to whole-genome sequence data from three strains of Saccharomyces cerevisiae2,4 and the sister species Saccharomyces paradoxus5. We observed a pattern of sequence variation among yeast strains in which ancestral recombination events lead to a mosaic of segments with shared genealogy. Based on sequence divergence and the inferred median size of shared segments (∼2,000 bp), we estimated that although any two strains have undergone approximately 16 million cell divisions since their last common ancestor, only 314 outcrossing events have occurred during this time (roughly one every 50,000 divisions). Local correlations in polymorphism rates indicate that linkage disequilibrium in yeast should extend over kilobases. Our results provide the initial foundation for population studies of association between genotype and phenotype in S. cerevisiae.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mortimer, R.K. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10, 403–409 (2000).
Goffeau, A. et al. Life with 6,000 genes. Science 274, 546–563–7 (1996).
Landry, C.R., Townsend, J.P., Hartl, D.L. & Cavalieri, D. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol. Ecol. 15, 575–591 (2006).
Gu, Z. et al. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102, 1092–1097 (2005).
Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003).
Aa, E., Townsend, J.P., Adams, R.I., Nielsen, K.M. & Taylor, J.W. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 702–715 (2006).
Fay, J.C. & Benavides, J.A. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1, 66–71 (2005).
Johnson, L.J. et al. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 166, 43–52 (2004).
Carvajal-Rodriguez, A., Crandall, K.A. & Posada, D. Recombination estimation under complex evolutionary models with the coalescent composite-likelihood method. Mol. Biol. Evol. 23, 817–827 (2006).
Hey, J. & Wakeley, J. A coalescent estimator of the population recombination rate. Genetics 145, 833–846 (1997).
Hudson, R.R. & Kaplan, N.L. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111, 147–164 (1985).
Enjalbert, J. & David, J.L. Inferring recent outcrossing rates using multilocus individual heterozygosity: application to evolving wheat populations. Genetics 156, 1973–1982 (2000).
Ronald, J., Tang, H. & Brem, R.B. Genome-wide evolutionary rates in laboratory and wild yeast. Genetics, published online 2 July 2006 (doi:10.1534/genetics.106.060863).
Baum, L.E. & Petrie, T. Statistical inference for probabilistic functions of finite state Markov chains. Ann. Math. Stat. 37, 1554–1563 (1966).
Przeworski, M., Hudson, R.R. & Di Rienzo, A. Adjusting the focus on human variation. Trends Genet. 16, 296–302 (2000).
Cherry, J.M. et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387, 67–73 (1997).
Liti, G., Peruffo, A., James, S.A., Roberts, I.N. & Louis, E.J. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22, 177–192 (2005).
Reich, D.E. et al. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32, 135–142 (2002).
McVean, G.A. A genealogical interpretation of linkage disequilibrium. Genetics 162, 987–991 (2002).
Mortimer, R. & Polsinelli, M. On the origins of wine yeast. Res. Microbiol. 150, 199–204 (1999).
Otto, S.P. & Lenormand, T. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3, 252–261 (2002).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Bray, N. & Pachter, L. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 14, 693–699 (2004).
Viterbi, A.J. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory 13, 260–269 (1967).
Hudson, R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Hein, J., Schierup, M.H. & Carsten, W. Gene Genealogies, Variation and Evolution (Oxford Univ. Press, New York, 2005).
Hudson, R.R., Slatkin, M. & Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992).
Acknowledgements
We thank D. Botstein, R. Brem, H. Coller, J. Ronald and especially M. Rockman for helpful discussions and comments on the manuscript. We thank the Broad Institute Fungal Genome Initiative, led by B. Birren, for generating the sequences of RM11-1a and S. paradoxus, and the Stanford Genome Technology Center, especially L. Steinmetz and R. Davis, for generating the sequence of YJM789. This work was supported by US National Institute of Mental Health grant R37 MH059520 and a James S. McDonnell Foundation Centennial Fellowship. SCP is supported by a grant from the Pew Charitable Trusts (award 2000-002558).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ruderfer, D., Pratt, S., Seidel, H. et al. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet 38, 1077–1081 (2006). https://doi.org/10.1038/ng1859
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1859
This article is cited by
-
Lessons from the Nakaseomyces: mating-type switching, DSB repair and evolution of Ho
Current Genetics (2021)
-
Mitochondrial-nuclear coadaptation revealed through mtDNA replacements in Saccharomyces cerevisiae
BMC Evolutionary Biology (2020)
-
Unlocking a signal of introgression from codons in Lachancea kluyveri using a mutation-selection model
BMC Evolutionary Biology (2020)
-
A yeast living ancestor reveals the origin of genomic introgressions
Nature (2020)
-
The fitness cost of mismatch repair mutators in Saccharomyces cerevisiae: partitioning the mutational load
Heredity (2020)