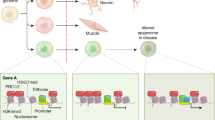
Abstract
Epigenetics generally refers to heritable changes in gene expression that are independent of nucleotide sequence. With complete genome sequences in hand, understanding the epigenetic control of genomes is the next step towards comprehending how the same DNA sequence gives rise to different cells, lineages and organs. Epigenetics also contributes to individual variation in normal biology and in disease states. The mouse provides a unique opportunity to understand how epigenetic differences contribute to both development and disease in a tractable mammalian system. Here we discuss current approaches and protocols used to study epigenetics in the mouse, including loss-of-function studies, mutagenesis screens, somatic cell nuclear transfer, genomics and proteomics.
This is a preview of subscription content, access via your institution
Access options



Similar content being viewed by others
References
Waddington, C. The epigenotype. Endeavor 1, 18–20 (1942).
Cowell, I.G. et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111, 22–36 (2002).
Sun, Z.W. & Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 (2002).
Costanzi, C. & Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Sarma, K. & Reinberg, D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6, 139–149 (2005).
Luger, K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 13, 127–135 (2003).
Zheng, C. & Hayes, J.J. Structures and interactions of the core histone tail domains. Biopolymers 68, 539–546 (2003).
Labrador, M. & Corces, V.G. Setting the boundaries of chromatin domains and nuclear organization. Cell 111, 151–154 (2002).
Goetze, S. et al. Performance of genomic bordering elements at predefined genomic loci. Mol. Cell. Biol. 25, 2260–2272 (2005).
Rousseaux, S. et al. Establishment of male-specific epigenetic information. Gene 345, 139–153 (2005).
Rangwala, S.H. & Richards, E.J. The value-added genome: building and maintaining genomic cytosine methylation landscapes. Curr. Opin. Genet. Dev. 14, 686–691 (2004).
Chen, T. & Li, E. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60, 55–89 (2004).
Hendrich, B. & Tweedie, S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19, 269–277 (2003).
Jorgensen, H.F. & Bird, A. MeCP2 and other methyl-CpG binding proteins. Ment. Retard. Dev. Disabil. Res. Rev. 8, 87–93 (2002).
Nakao, M. et al. Regulation of transcription and chromatin by methyl-CpG binding protein MBD1. Brain Dev. 23 Suppl 1, S174–S176 (2001).
Henikoff, S., Furuyama, T. & Ahmad, K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 20, 320–326 (2004).
Bell, A.C., West, A.G. & Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 (1999).
West, A.G., Huang, S., Gaszner, M., Litt, M.D. & Felsenfeld, G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16, 453–463 (2004).
Gebuhr, T.C., Bultman, S.J. & Magnuson, T. Pc-G/trx-G and the SWI/SNF connection: developmental gene regulation through chromatin remodeling. Genesis 26, 189–197 (2000).
Plath, K. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003).
Mager, J., Montgomery, N.D., de Villena, F.P. & Magnuson, T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33, 502–507 (2003).
Wang, J. et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28, 371–375 (2001).
Hernandez-Munoz, I. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102, 7635–7640 (2005).
Huttenhofer, A., Schattner, P. & Polacek, N. Non-coding RNAs: hope or hype? Trends Genet. 21, 289–297 (2005).
Roberts, E. & Quisenberry, J.H. Linkage of the genes for non-yellow (y) and pink-eye (p2) in the house mouse (Mus musculus). Am. Nat. 69, 181–183 (1935).
Vasicek, T.J. et al. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 147, 777–786 (1997).
Michaud, E.J. et al. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8, 1463–1472 (1994).
Rakyan, V. & Whitelaw, E. Transgenerational epigenetic inheritance. Curr. Biol. 13, R6 (2003).
Li, E., Bestor, T.H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Li, E., Beard, C. & Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993).
Sado, T. et al. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 225, 294–303 (2000).
Okano, M., Bell, D.W., Haber, D.A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Bourc'his, D., Xu, G.L., Lin, C.S., Bollman, B. & Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Guy, J., Hendrich, B., Holmes, M., Martin, J.E. & Bird, A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27, 322–326 (2001).
Hendrich, B., Guy, J., Ramsahoye, B., Wilson, V.A. & Bird, A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 15, 710–723 (2001).
Hutchins, A.S. et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell 10, 81–91 (2002).
Horike, S., Cai, S., Miyano, M., Cheng, J.F. & Kohwi-Shigematsu, T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37, 31–40 (2005).
Hemann, M.T. et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33, 396–400 (2003).
Kunath, T. et al. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 21, 559–561 (2003).
Fedoriw, A.M., Stein, P., Svoboda, P., Schultz, R.M. & Bartolomei, M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240 (2004).
Engel, N., West, A.G., Felsenfeld, G. & Bartolomei, M.S. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat. Genet. 36, 883–888 (2004).
Schoenherr, C.J., Levorse, J.M. & Tilghman, S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33, 66–69 (2003).
Russell, L.B. & Bangham, J.W. The paternal genome in mouse zygotes is less sensitive to ENU mutagenesis than the maternal genome. Mutat. Res. 248, 203–209 (1991).
Rinchik, E.M., Tonjes, R.R., Paul, D. & Potter, M.D. Molecular analysis of radiation-induced albino (c)-locus mutations that cause death at preimplantation stages of development. Genetics 135, 1107–1116 (1993).
Blewitt, M.E. et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102, 7629–7634 (2005).
Carpenter, A.E. & Sabatini, D.M. Systematic genome-wide screens of gene function. Nat. Rev. Genet. 5, 11–22 (2004).
Austin, C.P. et al. The knockout mouse project. Nat. Genet. 36, 921–924 (2004).
Wang, Y. et al. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 259, 3–17; discussion 17–21, 163–169 (2004).
Umlauf, D. et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36, 1296–1300 (2004).
Roh, T.Y., Ngau, W.C., Cui, K., Landsman, D. & Zhao, K. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 22, 1013–1016 (2004).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002).
Murrell, A., Heeson, S. & Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 (2004).
Bolzer, A. et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3, e157 (2005).
Eggan, K. et al. Mice cloned from olfactory sensory neurons. Nature 428, 44–49 (2004).
Li, J., Ishii, T., Feinstein, P. & Mombaerts, P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature 428, 393–399 (2004).
Mann, M.R. et al. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69, 902–914 (2003).
Boiani, M., Eckardt, S., Scholer, H.R. & McLaughlin, K.J. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16, 1209–1219 (2002).
Ogonuki, N. et al. Early death of mice cloned from somatic cells. Nat. Genet. 30, 253–254 (2002).
Eggan, K. et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98, 6209–6214 (2001).
Murrell, A., Rakyan, V.K. & Beck, S. From genome to epigenome. Hum. Mol. Genet. 14 Suppl 1, R3–R10 (2005).
Costello, J.F. et al. Aberrant CpG-island methylation has non-random and tumor-type-specific patterns. Nat. Genet. 24, 132–138 (2000).
Grunau, C., Hindermann, W. & Rosenthal, A. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum. Mol. Genet. 9, 2651–2663 (2000).
Rakyan, V.K. et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2, e405 (2004).
Song, F. et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl. Acad. Sci. USA 102, 3336–3341 (2005).
Buck, M.J. & Lieb, J.D. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83, 349–360 (2004).
Taverner, N.V., Smith, J.C. & Wardle, F.C. Identifying transcriptional targets. Genome Biol. 5, 210 (2004).
Bernstein, B.E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005).
Martens, J.H. et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812 (2005).
Mockler, T.C. & Ecker, J.R. Applications of DNA tiling arrays for whole-genome analysis. Genomics 85, 1–15 (2005).
Bertone, P. et al. Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246 (2004).
Cawley, S. et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509 (2004).
Yamada, K. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003).
Jiao, Y. et al. A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17, 1641–1657 (2005).
Acknowledgements
We thank S. Bultman and T. Magnuson for communication of unpublished results; D. Barlow and T. Jenuwein for discussion of European-based initiatives in epigenetics; and S. Reiner and members of the laboratory for critical reading of the manuscript. We apologize to those authors whose primary work could not be referenced owing to space limitations. This work was supported by the US National Institutes of Health and the Howard Hughes Medical Institute. J.M. is supported by the Damon Runyon Cancer Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mager, J., Bartolomei, M. Strategies for dissecting epigenetic mechanisms in the mouse. Nat Genet 37, 1194–1200 (2005). https://doi.org/10.1038/ng1664
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1664