Abstract
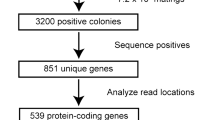
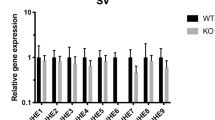
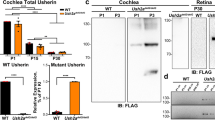
We identified a human mutation that causes dilated cardiomyopathy and heart failure preceded by sensorineural hearing loss (SNHL). Unlike previously described mutations causing dilated cardiomyopathy that affect structural proteins, this mutation deletes 4,846 bp of the human transcriptional coactivator gene EYA4. To elucidate the roles of eya4 in heart function, we studied zebrafish embryos injected with antisense morpholino oligonucleotides. Attenuated eya4 transcript levels produced morphologic and hemodynamic features of heart failure. To determine why previously described mutated EYA4 alleles1,2 cause SNHL without heart disease, we examined biochemical interactions of mutant Eya4 peptides. Eya4 peptides associated with SNHL, but not the shortened 193–amino acid peptide associated with dilated cardiomyopathy and SNHL, bound wild-type Eya4 and associated with Six proteins. These data define unrecognized and crucial roles for Eya4-Six–mediated transcriptional regulation in normal heart function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pfister, M. et al. A 4-bp insertion in the Eya-homologous region (EyaHR) of Eya4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 8, 607–611 (2002).
Wayne, S. et al. Mutations in the transcriptional activator Eya4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 10, 195–200 (2001).
Schönberger, J. et al. Dilated cardiomyopathy and sensorineural hearing loss: a heritable syndrome that maps to 6q23-24. Circulation 101, 1812–1818 (2000).
Bonini, N.M., Leiserson, W.M. & Benzer, S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379–395 (1993).
Borsani, G. et al. Eya4, a novel vertebrate gene related to Drosophila eyes absent. Hum. Mol. Genet. 8, 11–23 (1999).
Heanue, T.A. et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13, 3231–3243 (1999).
Ohto, H. et al. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19, 6815–6824 (1999).
Hanson, I.M. Mammalian homologues of the Drosophila eye specification genes. Semin. Cell Dev. Biol. 12, 475–484 (2001).
Bui, Q.T., Zimmerman, J.E., Liu, H. & Bonini, N.M. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155, 709–720 (2000).
Li, X. et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426, 247–254 (2003).
Rayapureddi, J.P. et al. Eyes absent represents a class of protein tyrosine phosphatases. Nature 426, 295–298 (2003).
Tootle, T.L. et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426, 299–302 (2003).
Xu, X. et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 30, 205–209 (2002).
Silver, S.J., Davies, E.L., Doyon, L. & Rebay, I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23, 5989–5999 (2003).
Jowett, T. Analysis of protein and gene expression. Methods Cell Biol. 59, 63–85 (1999).
Schwerte, T. & Pelster, B. Digital motion analysis as a tool for analysing the shape and performance of the circulatory system in transparent animals. J. Exp. Biol. 203, 1659–1669 (2000).
Acknowledgements
We thank the members of families MCE and MDB, without whose assistance this study would not have been possible, and H. Levy, B. Bruneau, B. McDonough, C. Duffy, J. Rivera-Feliciano, J.L. Galloway, D. Fatkin, H. Stäcker, R. Eavey, C. Halpin, D. Fox, V. VanderLaan and the Westshore Cardiology Group for helpful comments, technical expertise and support. This work was supported by grants from the Howard Hughes Medical Institutes (J.S., L.W., J.G.S. and C.E.S.), the National Institutes of Health (J.G.S. and C.E.S.), the German Research council (J.S.) and the Bugher foundation (J.S. and J.G.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Comparison of Eya4 protein sequences in zebrafish, mouse, and human. (PDF 75 kb)
Supplementary Fig. 2
Additional whole mount in situ hybridization of zebrafish Eya4 during embryogenesis. (PDF 10841 kb)
Rights and permissions
About this article
Cite this article
Schönberger, J., Wang, L., Shin, J. et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet 37, 418–422 (2005). https://doi.org/10.1038/ng1527
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1527
This article is cited by
-
Retinal determination gene networks: from biological functions to therapeutic strategies
Biomarker Research (2023)
-
Dilated cardiomyopathy caused by a pathogenic nucleotide variant in RBM20 in an Iranian family
BMC Medical Genomics (2022)
-
A combined genome-wide association and molecular study of age-related hearing loss in H. sapiens
BMC Medicine (2021)
-
Genome-first approach to rare EYA4 variants and cardio-auditory phenotypes in adults
Human Genetics (2021)
-
Insights into the pathophysiology of DFNA10 hearing loss associated with novel EYA4 variants
Scientific Reports (2020)