Abstract
We have determined the complete sequence of the mitochondrial DNA in the model plant species Arabidopsis thaliana, affording access to the first of its three genomes. The 366,924 nucleotides code for 57 identified genes, which cover only 10% of the genome. Introns in these genes add about 8%, open reading frames larger than 100 amino acids represent 10% of the genome, duplications account for 7%, remnants of retrotransposons of nuclear origin contribute 4% and integrated plastid sequences amount to 1% — leaving 60% of the genome unaccounted for. With the significant contribution of duplications, imported foreign DNA and the extensive background of apparently f unctionless sequences, the mosaic structure of the Arabidopsis thaliana mitochondrial genome features many aspects of size-relaxed nuclear genomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Meyerowitz,, E M. & Somerville, C.R. Arabidopsis (Cold Spring Harbor Laboratory Press, New York, 1994).
Blackbourn, H. & Ingram, J. Model progress with Arabidopsis. Trends Plant Sci. 1, 291–292 (1996).
Klein, M. et al. Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J. 6, 447–455 (1994).
Palmer, J.D. & Herbon, L.A. Unicircular structure of the Brassica hirta mitochondrial genome. Curr. Genet. 11, 565–570 (1987).
Ward, B.L., Anderson, R.S. & Bendich, A.J. The mitochondrial genome is large and variable in a family of plants. Cell 25, 793–803 (1981).
Oda, K. et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorphamitochondrial DNA: a primitive form of plant mitochondrial genome. Mol. Biol. 223, 1–7 (1992).
Brennicke, A., Grohmann, L., Hiesel, R., Knoop, V. & Schuster, W. The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBSLett. 325, 140–145 (1993).
Palmer, J.D. & Shields, C.R. Tripartite structure of the Brassica campestris mitochondrial genome. Nature 307, 411–415 (1984).
Covello, P.S. & Gray, M.W. RNA editing in plant mitochondria. Nature 341, 662–666 (1989).
Gualberto, J., Lamattina, L., Bonnard, G., Weil, J.-H. & Grienenberger, J.-M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341, 660–662 (1989).
Hiesel, R., Wissinger, B., Schuster, W. & Brennicke, A. RNA editing in plant mitochondria. Science 246, 1632–1634 (1989).
Chapdelaine, Y. & Bonen, L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a trans-splicing model for this gene-in-pieces. Cell 65, 465–472 (1991).
Wissinger, B., Schuster, W. & Brennicke, A. Trans-splicing in Oenothera mitochondria: nadl mRNAs are edited in exon and trans-splicing group II intron sequences. Cell. 65, 473–482 (1991).
Goodman, H.M., Ecker, J.R. & Dean, C. The genome of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92, 10831–10835 (1995).
Brandt, P., Sünkel, S., Unseld, M., Brennicke, A. & Knoop, V. The nad4L gene is encoded between exon c of nad5 and ORF25 in the Arabidopsis mitochondrial genome. Mol. Gen. Genet. 236, 33–38 (1992).
Binder, S. & Brennicke, A. Transcription initiation sites in mitochondria of Oenothera berteriana. J. Biol. Chem. 268, 7849–7855 (1993).
Fox, T.D. & Leaver, C.J., Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 26, 315–323 (1981).
Marechal-Drouard, L., Weil, J.-H. & Dietrich, A., RNAs and transfer RNA genes in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 13–32 (1993).
Knoop, V., Schuster, W., Wissinger, B. & Brennicke, A. Trans-splicing integrates an exon of 22 nucleotides into the nad5 mRNA in higher plant mitochondria. EMBOJ. 10, 3483–3493 (1991).
Knoop, V. & Brennicke, A. Group II introns in plant mitochondria - trans-splicing, RNA editing, evolution and promiscuity, in Plant Mitochondria (eds Brennicke, A. & Kuck, U.) 221–232 (VCH Chemie, Weinheim and New York, 1993).
Lippok, B., Brennicke, A. & Unseld, M. The rps4-gene is encoded upstream of the nad2-gene \nArabidopsismitochondria. Biol. Chem. 377, 251–257 (1996).
Ohta, E. et al. Group I introns in the liverwort mitochondrial genome: the gene coding for subunit 1 of cytochrome oxidase shares five intron positions with its fungal counterparts. Nucl. Acids Res. 5, 1297–1305 (1993).
Joyce, P.B.M. & Gray, M.W. Chloroplast-like transfer RNA genes expressed in wheat mitochondria. Nucl. Adds Res. 17, 5461–5476 (1989).
Knoop, V. et al. copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 142, 579–585 (1996).
Bendich, A.J. Reaching for the ring: the study of mitochondrial genome structure. Curr. Genet. 24, 279–290 (1993).
Knoop, V. & Brennicke, A. Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J. Mol. Evol. 39, 144–150 (1994).
Suplick, K., Morrisey, J. & Vaidya, A.B. Complex transcription from the extrachromosomal DNA encoding mitochondrial functions of Plasmodium yoelii. Mol. Cell. Biol. 10, 6381–6388 (1990).
Gray, M.W. & Boer, P.H. Organization and expression of algal (Chlamydomonas reinhardtii)mitochondrial DNAPhil. Trans. R. Soc. Lond. B 319, 135–147 (1988).
Blanchard, J.L. & Schmidt, G.W. Pervasive migration of organellar DNA to the nucleus in plants. J. Mol. Evol. 41, 397–406 (1995).
Michaelis, G., Vahrenholz, C. & Pratje, E. Mitochondrial DNA of Chlamydomonas reinhardtii: The gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol. Gen. Genet. 223, 211–216 (1990).
Brenner, S.E., Hubbard, T., Murzin, A. & Chothia, C. Gene duplications in Haemophilus influenza. Nature 378, 140 (1995).
Levings III., C.S. The Texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science 250, 942–947 (1990).
Knoop, V., Kloska, S. & Brennicke, A. On the identification of group II introns in nucleotide sequence data. J. Mol. Biol. 242, 389–396 (1994).
Lang, B.F., Goff, U. & Gray, M.W. A 5 S rRNA gene is present in the mitochondrial genome of the protist Redimonas americana but is absent from red algal mitochondrial DNA. J. Mol. Biol. 261, 607–613 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Unseld, M., Marienfeld, J., Brandt, P. et al. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15, 57–61 (1997). https://doi.org/10.1038/ng0197-57
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0197-57
This article is cited by
-
Xenografted human microglia display diverse transcriptomic states in response to Alzheimer’s disease-related amyloid-β pathology
Nature Neuroscience (2024)
-
TALE-based organellar genome editing and gene expression in plants
Plant Cell Reports (2024)
-
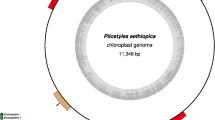
Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca
BMC Genomics (2021)
-
Comparative analysis of nuclear, chloroplast, and mitochondrial genomes of watermelon and melon provides evidence of gene transfer
Scientific Reports (2021)
-
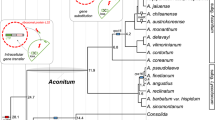
Plant Mitochondria are a Riddle Wrapped in a Mystery Inside an Enigma
Journal of Molecular Evolution (2021)