Abstract
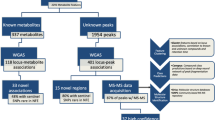
We present a genome-wide association study of metabolic traits in human urine, designed to investigate the detoxification capacity of the human body. Using NMR spectroscopy, we tested for associations between 59 metabolites in urine from 862 male participants in the population-based SHIP study. We replicated the results using 1,039 additional samples of the same study, including a 5-year follow-up, and 992 samples from the independent KORA study. We report five loci with joint P values of association from 3.2 × 10−19 to 2.1 × 10−182. Variants at three of these loci have previously been linked with important clinical outcomes: SLC7A9 is a risk locus for chronic kidney disease, NAT2 for coronary artery disease and genotype-dependent response to drug toxicity, and SLC6A20 for iminoglycinuria. Moreover, we identify rs37369 in AGXT2 as the genetic basis of hyper-β-aminoisobutyric aciduria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gieger, C. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4, e1000282 (2008).
Illig, T. et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42, 137–141 (2010).
Tanaka, T. et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5, e1000338 (2009).
Hicks, A.A. et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 5, e1000672 (2009).
Weljie, A.M., Newton, J., Mercier, P., Carlson, E. & Slupsky, C.M. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 78, 4430–4442 (2006).
Altmaier, E. et al. Bioinformatics analysis of targeted metabolomics—uncovering old and new tales of diabetic mice under medication. Endocrinology 149, 3478–3489 (2008).
Suhre, K. et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE 5, e13953 (2010).
Crumpler, H.R., Dent, C.E., Harris, H. & Westall, R.G. β-Aminoisobutyric acid (α-methyl-β-alanine); a new amino-acid obtained from human urine. Nature 167, 307–308 (1951).
Harris, H. Family studies on the urinary excretion of beta-aminoisobutyric acid. Ann. Eugen. 18, 43–49 (1953).
Yanai, J., Kakimoto, Y., Tsujio, T. & Sano, I. Genetic study of beta-aminoisobutyric acid excretion by Japanese. Am. J. Hum. Genet. 21, 115–132 (1969).
Kakimoto, Y., Taniguchi, K. & Sano, I. D-beta-aminoisobutyrate:pyruvate aminotransferase in mammalian liver and excretion of beta-aminoisobutyrate by man. J. Biol. Chem. 244, 335–340 (1969).
van Kuilenburg, A.B. et al. β-Ureidopropionase deficiency: an inborn error of pyrimidine degradation associated with neurological abnormalities. Hum. Mol. Genet. 13, 2793–2801 (2004).
Scriver, C.R. & Perry, T.L. Disorders of omega-amino acids in free and peptide-linked forms. in The Metabolic Basis of Inherited Disease Vol. I (eds. Scriver, C.R., Beaudet, A.L., Sly, W.S. & Valle, D.) 755–771 (McGraw-Hill, New York, 1989).
Rodionov, R.N., Murry, D.J., Vaulman, S.F., Stevens, J.W. & Lentz, S.R. Human alanine-glyoxylate aminotransferase 2 lowers asymmetric dimethylarginine and protects from inhibition of nitric oxide production. J. Biol. Chem. 285, 5385–5391 (2010).
Kayrak, M. et al. Association between exaggerated blood pressure response to exercise and serum asymmetric dimethylarginine levels. Circ. J. 74, 1135–1141 (2010).
Wilson Tang, W.H. et al. Differential effects of arginine methylation on diastolic dysfunction and disease progression in patients with chronic systolic heart failure. Eur. Heart J. 29, 2506–2513 (2008).
Vallance, P., Leone, A., Calver, A., Collier, J. & Moncada, S. Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 339, 572–575 (1992).
Fliser, D. et al. Asymmetric dimethylarginine and progression of chronic kidney disease: the mild to moderate kidney disease study. J. Am. Soc. Nephrol. 16, 2456–2461 (2005).
Ravani, P. et al. Asymmetrical dimethylarginine predicts progression to dialysis and death in patients with chronic kidney disease: a competing risks modeling approach. J. Am. Soc. Nephrol. 16, 2449–2455 (2005).
Leiper, J. et al. Disruption of methylarginine metabolism impairs vascular homeostasis. Nat. Med. 13, 198–203 (2007).
Sanderson, S., Salanti, G. & Higgins, J. Joint effects of the N-acetyltransferase 1 and 2 (NAT1 and NAT2) genes and smoking on bladder carcinogenesis: a literature-based systematic HuGE review and evidence synthesis. Am. J. Epidemiol. 166, 741–751 (2007).
Deeken, J.F. et al. A pharmacogenetic study of docetaxel and thalidomide in patients with castration-resistant prostate cancer using the DMET genotyping platform. Pharmacogenomics J. 10, 191–199 (2010).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Daly, A.K. Drug-induced liver injury: past, present and future. Pharmacogenomics 11, 607–611 (2010).
Soranzo, N. et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 41, 1182–1190 (2009).
Scott, C.R. The genetic tyrosinemias. Am. J. Med. Genet. C. Semin. Med. Genet. 142C, 121–126 (2006).
Mattoo, A. & Goldfarb, D.S. Cystinuria. Semin. Nephrol. 28, 181–191 (2008).
Evan, A.P. et al. Renal crystal deposits and histopathology in patients with cystine stones. Kidney Int. 69, 2227–2235 (2006).
Köttgen, A. et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42, 376–384 (2010).
Chambers, J.C. et al. Genetic loci influencing kidney function and chronic kidney disease. Nat. Genet. 42, 373–375 (2010).
Takanaga, H., Mackenzie, B., Suzuki, Y. & Hediger, M.A. Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system imino. J. Biol. Chem. 280, 8974–8984 (2005).
Bröer, S. et al. Iminoglycinuria and hyperglycinuria are discrete human phenotypes resulting from complex mutations in proline and glycine transporters. J. Clin. Invest. 118, 3881–3892 (2008).
Volzke, H. et al. Cohort profile: the Study of Health in Pomerania. Int. J. Epidemiol. 40, 294–307 (2011).
John, U. et al. Study of Health In Pomerania (SHIP): a health examination survey in an east German region: objectives and design. Soz. Praventivmed. 46, 186–194 (2001).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Acknowledgements
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the German Federal Ministry of Education and Research (BMBF; grants 01ZZ9603, 01ZZ0103 and 01ZZ0403), the German Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg–West Pomerania. Genome-wide studies were supported by BMBF (grant 03ZIK012) and a joint grant from Siemens Healthcare and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Siemens 'Center of Knowledge Interchange' program. NMR studies were supported by Bruker BioSpin. This study was also supported in part by a grant from BMBF to the German Center for Diabetes Research (DZD e.V.), and by the Genomics of Lipid-associated Disorders project of the Austrian Genome Research Programme. This work is also part of the research project Greifswald Approach to Individualized Medicine (GANI_MED). The GANI_MED consortium is funded by the BMBF and the Ministry of Cultural Affairs of the Federal State of Mecklenburg–West Pomerania (03IS2061A). The KORA research platform and the 'Monitoring trends and determinants on cardiovascular diseases' (MONICA) Augsburg studies were initiated and financed by the Helmholtz Zentrum München–National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFNPlus 01GS0823). Part of this research was supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. J.R. is supported by Deutsche Forschungsgemeinschaft Graduiertenkolleg 'GRK 1563, Regulation and Evolution of Cellular Systems' (RECESS). Computing resources have been provided by the Leibniz Supercomputing Centre of the Bavarian Academy of Sciences and Humanities (HLRB project h1231) and the DEISA Extreme Computing Initiative (project PHAGEDA). We thank P. Lichtner, G. Eckstein, G. Fischer, T. Strom and all other members of the Helmholtz Zentrum München genotyping staff for generating the KORA SNP data set, as well as all field staff members involved in the MONICA and KORA Augsburg studies. The KORA group consists of H.E. Wichmann (speaker), A. Peters, C. Meisinger, T. Illig, R. Holle, J. John and their co-workers, who are responsible for the design and conduct of the KORA studies. We thank all participants in the SHIP and KORA studies for donating their blood, urine and time.
Author information
Authors and Affiliations
Contributions
K.S., H.W. and M.N. conceived and designed the experiments. H.W., N.F., R.H., K.M., C.W., A.K. and U.V. performed the experiments. K.S. and J.R. performed statistical analysis. K.S., H.W., J.R., F.K., N.F., D.C., A.T., C.G. and W.R.-M. analyzed the data. W.H., T.K., S.B.F., H.V. and R.B. designed and conducted the SHIP study. C.M, H.-E.W. and T.I. designed and conducted the KORA study. C.M., H.-E.W., T.I., H.K.K. and M.N. contributed reagents, materials and analysis tools. K.S., H.W., F.K., C.G. and M.N. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
D.C. is an employee of Chenomx Inc., which sells the software suite used for the NMR analysis in this study.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Note. (PDF 1196 kb)
Supplementary Table 1
MS-Excel data file, providing 15,475 associations that are significant at the 5% level after correcting for testing 1,720 metabolic traits at a single locus (p < 2.9×10−5) and that have a p-gain > 59 in the case of ratios (XLS 8487 kb)
Supplementary Table 2
MS-Excel data file providing associations based on 1000-Genomes data imputed genotypes at the five loci reported in Table 1 (XLS 11483 kb)
Rights and permissions
About this article
Cite this article
Suhre, K., Wallaschofski, H., Raffler, J. et al. A genome-wide association study of metabolic traits in human urine. Nat Genet 43, 565–569 (2011). https://doi.org/10.1038/ng.837
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.837
This article is cited by
-
Genetic loci of beta-aminoisobutyric acid are associated with aging-related mild cognitive impairment
Translational Psychiatry (2023)
-
Effects of AGXT2 variants on blood pressure and blood sugar among 750 older Japanese subjects recruited by the complete enumeration survey method
BMC Genomics (2021)
-
Rare genetic variants affecting urine metabolite levels link population variation to inborn errors of metabolism
Nature Communications (2021)
-
Urinary metabolic phenotyping for Alzheimer’s disease
Scientific Reports (2020)
-
Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans
Nature Genetics (2020)