Abstract
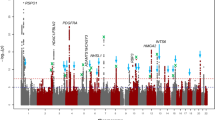
Refractive errors are the most common ocular disorders worldwide and may lead to blindness. Although this trait is highly heritable, identification of susceptibility genes has been challenging. We conducted a genome-wide association study for refractive error in 5,328 individuals from a Dutch population-based study with replication in four independent cohorts (combined 10,280 individuals in the replication stage). We identified a significant association at chromosome 15q14 (rs634990, P = 2.21 × 10−14). The odds ratio of myopia compared to hyperopia for the minor allele (minor allele frequency = 0.47) was 1.41 (95% CI 1.16–1.70) for individuals heterozygous for the allele and 1.83 (95% CI 1.42–2.36) for individuals homozygous for the allele. The associated locus is near two genes that are expressed in the retina, GJD2 and ACTC1, and appears to harbor regulatory elements which may influence transcription of these genes. Our data suggest that common variants at 15q14 influence susceptibility for refractive errors in the general population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Bourne, R.R., Dineen, B.P., Ali, S.M., Noorul Huq, D.M. & Johnson, G.J. Prevalence of refractive error in Bangladeshi adults: results of the National Blindness and Low Vision Survey of Bangladesh. Ophthalmology 111, 1150–1160 (2004).
Dandona, R. et al. Population-based assessment of refractive error in India: the Andhra Pradesh eye disease study. Clin. Experiment. Ophthalmol. 30, 84–93 (2002).
Kempen, J.H. et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch. Ophthalmol. 122, 495–505 (2004).
Sawada, A. et al. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology 115, 363–370 (2008).
Vitale, S., Ellwein, L., Cotch, M.F., Ferris, F.L. III & Sperduto, R. Prevalence of refractive error in the United States, 1999–2004. Arch. Ophthalmol. 126, 1111–1119 (2008).
McBrien, N.A. & Gentle, A. Role of the sclera in the development and pathological complications of myopia. Prog. Retin. Eye Res. 22, 307–338 (2003).
Saw, S.M. et al. How blinding is pathological myopia? Br. J. Ophthalmol. 90, 525–526 (2006).
Curtin, B.J. & Karlin, D.B. Axial length measurements and fundus changes of the myopic eye. Am. J. Ophthalmol. 1, 42–53 (1971).
Saw, S.M., Gazzard, G., Shih-Yen, E.C. & Chua, W.H. Myopia and associated pathological complications. Ophthalmic Physiol. Opt. 25, 381–391 (2005).
Tano, Y. et al. Pathologic myopia: where are we now? Am. J. Ophthalmol. 134, 645–660 (2002).
Young, T.L. et al. Molecular genetics of human myopia: an update. Optom. Vis. Sci. 86, E8–E22 (2009).
Dirani, M. et al. Outdoor activity and myopia in Singapore teenage children. Br. J. Ophthalmol. 93, 997–1000 (2009).
McBrien, N.A. et al. Myopia: recent advances in molecular studies; prevalence, progression and risk factors; emmetropization; therapies; optical links; peripheral refraction; sclera and ocular growth; signalling cascades; and animal models. Optom. Vis. Sci. published online, doi:10.1097/01.opx.0000344146.84135.68 (19 December 2008).
Saw, S.M., Hong, C.Y., Chia, K.S., Stone, R.A. & Tan, D. Nearwork and myopia in young children. Lancet 357, 390 (2001).
Young, T.L., Metlapally, R. & Shay, A.E. Complex trait genetics of refractive error. Arch. Ophthalmol. 125, 38–48 (2007).
Eystathioy, T., Jakymiw, A., Fujita, D.J., Fritzler, M.J. & Chan, E.K. Human autoantibodies to a novel Golgi protein golgin-67: high similarity with golgin-95/gm 130 autoantigen. J. Autoimmun. 14, 179–187 (2000).
Jobling, A.I., Gentle, A., Metlapally, R., McGowan, B.J. & McBrien, N.A. Regulation of scleral cell contraction by transforming growth factor-beta and stress: competing roles in myopic eye growth. J. Biol. Chem. 284, 2072–2079 (2009).
Kihara, A.H. et al. Connexin36, an essential element in the rod pathway, is highly expressed in the essentially rodless retina of Gallus gallus. J. Comp. Neurol. 512, 651–663 (2009).
Deans, M.R., Volgyi, B., Goodenough, D.A., Bloomfield, S.A. & Paul, D.L. Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron 36, 703–712 (2002).
Striedinger, K. et al. Loss of connexin36 increases retinal cell vulnerability to secondary cell loss. Eur. J. Neurosci. 22, 605–616 (2005).
Güldenagel, M. et al. Visual transmission deficits in mice with targeted disruption of the gap junction gene connexin36. J. Neurosci. 21, 6036–6044 (2001).
Rong, P. et al. Disruption of Gja8 (a8 connexin) in mice leads to microphthalmia associated with retardation of lens growth and lens fiber maturation. Development 129, 167–174 (2002).
White, T.W. Targeted ablation of Connexin50 in mice results in microphthalmia and zonular pulverulent cataracts. J. Cell Biol. 143, 815–825 (1998).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Heintzman, N.D. & Ren, B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541–549 (2009).
Delaloy, C. et al. Deletion of WNK1 first intron results in misregulation of both isoforms in renal and extrarenal tissues. Hypertension 52, 1149–1154 (2008).
Mihaly, J. et al. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133, 2983–2993 (2006).
Hysi, P.G., Young, T.L. et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25 which contains the RASGRF1 gene. Nat. Genet. advance online publication, doi:10.1038/ng.664 (12 September 2010).
Jones, C. & Moses, K. Cell-cycle regulation and cell-type specification in the developing Drosophila compound eye. Semin. Cell Dev. Biol. 15, 75–81 (2004).
Fernández-Medarde, A. et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J. Neurochem. 110, 641–652 (2009).
Lettre, G. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 40, 584–591 (2008).
Hofman, A. et al. The Rotterdam Study: 2010 objectives and design update. Eur. J. Epidemiol. 24, 553–572 (2009).
Spector, T.D. & MacGregor, A.J. The St. Thomas' UK Adult Twin Registry. Twin Res. 5, 440–443 (2002).
Estrada, K. et al. GRIMP: A web- and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics 25, 2750–2752 (2009).
Aulchenko, Y.S., Ripke, S., Isaacs, A. & van Duijn, C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Booij, J.C. et al. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics 20, 10–164 (2009).
Van Soest, S.S. et al. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol. Vis. 10, 1608–1617 (2007).
Acknowledgements
Major funding of the work performed in The Netherlands came from the Netherlands Organisation of Scientific Research (NWO); Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands; Netherlands Organization for Health Research and Development (ZonMw); UitZicht; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); the Municipality of Rotterdam; the Netherlands Genomics Initiative (NGI)/NWO; Center for Medical Systems Biology of NGI; Lijf en Leven; M.D. Fonds; H. Stichting; Oogfonds Nederland; Stichting Nederlands Oogheelkundig Onderzoek; Swart van Essen; Bevordering van Volkskracht; Blindenhulp; Landelijke Stichting voor Blinden en Slechtzienden; Rotterdamse Vereniging voor Blindenbelangen; OOG; Algemene Nederlandse Vereniging ter Voorkoming van Blindheid; the Rotterdam Eye Hospital Research Foundation; Laméris Ootech; Topcon Europe; and Heidelberg Engineering. We thank A. Hooghart, C. Brussee, R. Bernaerts-Biskop, P. van Hilten, P. Arp, M. Jhamai, M. Moorhouse, J. Vergeer, M. Verkerk, S. Bervoets and P. van der Spek for help in execution of the study.
TwinsUK acknowledges the Wellcome Trust, the European Union MyEuropia Marie Curie Research Training Network, Guide Dogs for the Blind Association, the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007-2013), European Network of Genetic and Genomic Epidemiology (ENGAGE) project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), Biotechnology and Biological Sciences Research Council (G20234), Department of Health via US National Institutes of Health, (National Eye Institute grant RO1EY018246), and the Center for Inherited Disease Research. TwinsUK thanks G. Surdulescu, L. Peltonen, P. Deloukas, M. Lathrop, D. Goldstein, A. Palotie and C. Day for help in execution of the study and analyses.
Author information
Authors and Affiliations
Contributions
A.M.S., V.J.M.V. and C.C.W.K. performed analyses and drafted the manuscript. C.M.v.D., B.A.O., F.R., A.G.U., A.H., P.T.V.M.d.J., J.R.V. and C.C.W.K. designed the study and obtained funding. D.D.G.D., L.M.v.K., L.H., W.D.R., M.C., R.K., J.J.M.W.-A., T.G.M.F.G., F.C.C.R. and S.M.A.S. helped in data collection. A.J.M.H.V., M.K.I., N.A., M.S., Y.S.A., A.A.B.B., A.A.L.J.v.O. and A.I. participated in the genetic analyses. P.G.H., T.L.Y., D.A.M., T.D.S. and C.J.H. were responsible for data from the TwinsUK study. M.K.I., R.W.A.M.K., G.v.R., P.G.H., C.J.H., C.M.v.D., A.J.M.H.V., B.A.O., J.R.V. and A.A.B.B. critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5 and Supplementary Figues 1–3. (PDF 449 kb)
Rights and permissions
About this article
Cite this article
Solouki, A., Verhoeven, V., van Duijn, C. et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet 42, 897–901 (2010). https://doi.org/10.1038/ng.663
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.663
This article is cited by
-
Vitamin D and myopia: a review
International Ophthalmology (2024)
-
The Role of Retinal Dysfunction in Myopia Development
Cellular and Molecular Neurobiology (2023)
-
Loss of Gap Junction Delta-2 (GJD2) gene orthologs leads to refractive error in zebrafish
Communications Biology (2021)
-
RNA-seq and GSEA identifies suppression of ligand-gated chloride efflux channels as the major gene pathway contributing to form deprivation myopia
Scientific Reports (2021)
-
Association of 5p15.2 and 15q14 with high myopia in Tujia and Miao Chinese populations
BMC Ophthalmology (2020)