Abstract
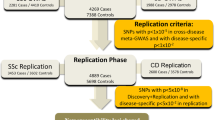
To identify susceptibility loci for ankylosing spondylitis, we undertook a genome-wide association study in 2,053 unrelated ankylosing spondylitis cases among people of European descent and 5,140 ethnically matched controls, with replication in an independent cohort of 898 ankylosing spondylitis cases and 1,518 controls. Cases were genotyped with Illumina HumHap370 genotyping chips. In addition to strong association with the major histocompatibility complex (MHC; P < 10−800), we found association with SNPs in two gene deserts at 2p15 (rs10865331; combined P = 1.9 × 10−19) and 21q22 (rs2242944; P = 8.3 × 10−20), as well as in the genes ANTXR2 (rs4333130; P = 9.3 × 10−8) and IL1R2 (rs2310173; P = 4.8 × 10−7). We also replicated previously reported associations at IL23R (rs11209026; P = 9.1 × 10−14) and ERAP1 (rs27434; P = 5.3 × 10−12). This study reports four genetic loci associated with ankylosing spondylitis risk and identifies a major role for the interleukin (IL)-23 and IL-1 cytokine pathways in disease susceptibility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Braun, J. et al. Prevalence of spondylarthropathies in HLA-B27 positive and negative blood donors. Arthritis Rheum. 41, 58–67 (1998).
Calin, A., Marder, A., Becks, E. & Burns, T. Genetic differences between B27 positive patients with ankylosing spondylitis and B27 positive healthy controls. Arthritis Rheum. 26, 1460–1464 (1983).
van der Linden, S., Valkenburg, H. & Cats, A. The risk of developing ankylosing spondylitis in HLA-B27 positive individuals: a family and population study. Br. J. Rheumatol. 22, 18–19 (1983).
Brown, M.A. et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 40, 1823–1828 (1997).
Brown, M.A., Laval, S.H., Brophy, S. & Calin, A. Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann. Rheum. Dis. 59, 883–886 (2000).
WTCCC & TASC. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Laval, S.H. et al. Whole-genome screening in ankylosing spondylitis: evidence of non-MHC genetic-susceptibility loci. Am. J. Hum. Genet. 68, 918–926 (2001).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Goto, Y., Tanji, H., Hattori, A. & Tsujimoto, M. Glutamine-181 is crucial in the enzymatic activity and substrate specificity of human endoplasmic-reticulum aminopeptidase-1. Biochem. J. 416, 109–116 (2008).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Kugathasan, S. et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 40, 1211–1215 (2008).
Mielants, H. et al. The evolution of spondyloarthropathies in relation to gut histology. II. Histological aspects. J. Rheumatol. 22, 2273–2278 (1995).
Taft, R.J. et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578 (2009).
Cui, X., Rouhani, F.N., Hawari, F. & Levine, S.J. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J. Immunol. 171, 6814–6819 (2003).
Cui, X. et al. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J. Clin. Invest. 110, 515–526 (2002).
Lappalainen, M. et al. Association of IL23R, TNFRSF1A, and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm. Bowel Dis. 14, 1118–1124 (2008).
Waschke, K.A. et al. Tumor necrosis factor receptor gene polymorphisms in Crohn′s disease: association with clinical phenotypes. Am. J. Gastroenterol. 100, 1126–1133 (2005).
Armaka, M. et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205, 331–337 (2008).
van der Linden, S., Valkenburg, H.A. & Cats, A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 27, 361–368 (1984).
Timpson, N.J. et al. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum. Mol. Genet. 18, 1510–1517 (2009).
Price, A.L. et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83, 132–135 author reply 135–139 (2008).
Kiiveri, H. A Bayesian approach to variable selection when the number of variables is very large. in Science and Statistics: A Festschrift for Terry Speed vol. 40 (ed. Goldstein, D.R.) 127–143 (Institute of Mathematical Statistics. Beachwood, Ohio, USA, 2003).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2009).
Lusa, L., Gentleman, R. & Ruschhaupt, M. GeneMeta: MetaAnalysis for High Throughput Experiments. R package version 1.16.0. (2008).
Acknowledgements
We would like to thank all participants, with and without ankylosing spondylitis, who provided the case and control DNA and clinical information necessary for this study. We are also very grateful for the invaluable support received from the National Ankylosing Spondylitis Society (UK) and Spondyloarthritis Association of America in subject recruitment. We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole Avon Longitudinal Study of Parents And Children (ALSPAC) team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. This study was funded by US National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants P01-052915 and R01-AR046208. Funding was also received from the University of Texas at Houston Clinical and Translational Science Award grant UL1RR024188, Cedars-Sinai General Clinical Research Center grant MO1-RR00425, Intramural Research Program of NIAMS/National Institutes of Health, and Rebecca Cooper Foundation (Australia). This study was funded in part by the Arthritis Research Campaign (UK), by the Wellcome Trust (grant number 076113) and by the Oxford Radcliffe Hospital Biomedical Research Centre ankylosing spondylitis chronic disease cohort (theme code A91202). M.A.B. is funded by the National Health and Medical Research Council (Australia). The UK Medical Research Council, the Wellcome Trust and the University of Bristol provide core support for ALSPAC.
Author information
Authors and Affiliations
Consortia
Contributions
Case assessment and recruitment: L.A.B., M.A.B., J.C.D., L.D., C.F., R.D.I., W.P.M., R.M., E.P., J.D.R., L.S., M.A.S., M.M.W., M.H.W. and B.P.W. Sample processing and genotyping: L.H.A., P.D., T.D., A.D., E.L.D., J.H., D.H., R.J., T.K., R.M., J.J.P., E.P., K.P., A.-M.S., J.T. and X.Z. Gene expression studies: R.D., E.G. and G.P.T. Data analysis: M.A.B., P.D., D.M.E. and P.L. Study design and management committee: M.A.B., J.D.R., M.M.W., M.H.W. and B.P.W. Manuscript preparation: M.A.B., D.M.E., P.L., J.D.R., M.A.S., M.H.W. and B.P.W. ALSPAC control data: P.D., V.K., L.P., S.M.R. and P.W.
Corresponding authors
Ethics declarations
Competing interests
M.A.B. is an employee of the University of Queensland, which has applied for patents relating to genes discovered in this study.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 731 kb)
Rights and permissions
About this article
Cite this article
The Australo-Anglo-American Spondyloarthritis Consortium (TASC). Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet 42, 123–127 (2010). https://doi.org/10.1038/ng.513
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.513
This article is cited by
-
Zur Rolle von HLA-B27 in der Pathogenese und Diagnostik der axialen Spondyloarthritis
Zeitschrift für Rheumatologie (2024)
-
Frontiers of ankylosing spondylitis research: an analysis from the top 100 most influential articles in the field
Clinical and Experimental Medicine (2023)
-
Integrative blood-derived epigenetic and transcriptomic analysis reveals the potential regulatory role of DNA methylation in ankylosing spondylitis
Arthritis Research & Therapy (2022)
-
A new and spontaneous animal model for ankylosing spondylitis is found in cynomolgus monkeys
Arthritis Research & Therapy (2022)
-
CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity
Nature Communications (2022)