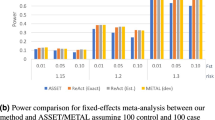
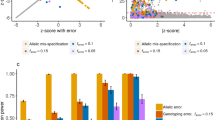
Abstract
Aggregate results from genome-wide association studies (GWAS)1,2,3, such as genotype frequencies for cases and controls, were until recently often made available on public websites4,5 because they were thought to disclose negligible information concerning an individual's participation in a study. Homer et al.6 recently suggested that a method for forensic detection of an individual's contribution to an admixed DNA sample could be applied to aggregate GWAS data. Using a likelihood-based statistical framework, we developed an improved statistic that uses genotype frequencies and individual genotypes to infer whether a specific individual or any close relatives participated in the GWAS and, if so, what the participant's phenotype status is. Our statistic compares the logarithm of genotype frequencies, in contrast to that of Homer et al.6, which is based on differences in either SNP probe intensity or allele frequencies. We derive the theoretical power of our test statistics and explore the empirical performance in scenarios with varying numbers of randomly chosen or top-associated SNPs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Manolio, T.A., Brooks, L.D. & Collins, F.S.A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008).
McCarthy, M.I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 (2008).
Wang, W.Y., Barratt, B.J., Clayton, D.G. & Todd, J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 6, 109–118 (2005).
Mailman, M.D. et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Homer, N. et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 4, e1000167 (2008).
Author information
Authors and Affiliations
Contributions
K.B.J. and N.C. devised the statistical methods; K.B.J. implemented these methods in software and applied them to simulated and empirical data; K.B.J., M.Y. and S.J.C. drafted the article; S.W. and P.K. made important suggestions to the analytic plan and aided in the interpretation of the results; D.C. and J.P. aided in the study design and verification of methodology; D.J.H., T.A.M., M.T., R.N.H. and G.D.T. participated in revising the manuscript and made important intellectual contributions. All the authors and reviewed and approved the final manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 545 kb)
Rights and permissions
About this article
Cite this article
Jacobs, K., Yeager, M., Wacholder, S. et al. A new statistic and its power to infer membership in a genome-wide association study using genotype frequencies. Nat Genet 41, 1253–1257 (2009). https://doi.org/10.1038/ng.455
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.455
This article is cited by
-
Routes for breaching and protecting genetic privacy
Nature Reviews Genetics (2014)
-
Methods for the de-identification of electronic health records for genomic research
Genome Medicine (2011)
-
Capture and release
Nature Genetics (2011)
-
Multiple common variants for celiac disease influencing immune gene expression
Nature Genetics (2010)
-
Not so lost in the genetic crowd
Nature Genetics (2009)