Abstract
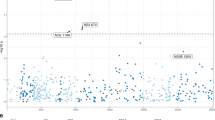
Hepatitis C virus (HCV) infects 3% of the world's population. Treatment of chronic HCV consists of a combination of PEGylated interferon-α (PEG-IFN-α) and ribavirin (RBV). To identify genetic variants associated with HCV treatment response, we conducted a genome-wide association study of sustained virological response (SVR) to PEG-IFN-α/RBV combination therapy in 293 Australian individuals with genotype 1 chronic hepatitis C, with validation in an independent replication cohort consisting of 555 individuals. We report an association to SVR within the gene region encoding interleukin 28B (IL28B, also called IFNλ3; rs8099917 combined P = 9.25 × 10−9, OR = 1.98, 95% CI = 1.57–2.52). IL28B contributes to viral resistance and is known to be upregulated by interferons and by RNA virus infection. These data suggest that host genetics may be useful for the prediction of drug response, and they also support the investigation of the role of IL28B in the treatment of HCV and in other diseases treated with IFN-α.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
World Health Organization (WHO). Hepatitis C. Fact Sheet No. 164. Revised October 2000 http://www.who.int/mediacentre/factsheets/fs164/en/ (2000).
Micallef, J.M., Kaldor, J.M. & Dore, G.J. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J. Viral Hepat. 13, 34–41 (2006).
Hoofnagle, J.H. Course and outcome of hepatitis C. Hepatology. 36, S21–S29 (2002).
Thomas, D.L. & Seeff, L.B. Natural history of hepatitis C. Clin. Liver Dis. 9, 383–398, vi (2005).
Di Bisceglie, A.M. & Hoofnagle, J.H. Optimal therapy of hepatitis C. Hepatology. 36, S121–S127 (2002).
David, M. Signal transduction by type I interferons. Biotechniques (Oct), S58–S65 (2002).
Der, S.D., Zhou, A., Williams, B.R. & Silverman, R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95, 15623–15628 (1998).
de Veer, M.J. et al. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69, 912–920 (2001).
Taylor, M.W. et al. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J. Interferon Cytokine Res. 24, 107–118 (2004).
Tan, H. et al. Global transcriptional profiling combination of type I and type II demonstrates the interferon enhances antiviral and immune responses at clinically relevant doses. J. Interferon Cytokine Res. 25, 632–649 (2005).
Taylor, M.W. et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from non-responders to antiviral therapy. J. Virol. 81, 3391–3401 (2007).
Fried, M.W. et al. A multicenter, randomized trial of daily high-dose interferon-alfa 2b for the treatment of chronic hepatitis c: pretreatment stratification by viral burden and genotype. Am. J. Gastroenterol. 95, 3225–3229 (2000).
Manns, M.P. et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomized trial. Lancet 358, 958–965 (2001).
Fried, M.W. et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347, 975–982 (2002).
Hadziyannis, S.J. et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140, 346–355 (2004).
Zeuzem, S. et al. Expert opinion on the treatment of patients with chronic hepatitis C. J. Viral Hepat. 16, 75–90 (2009).
Kraft, P. et al. Beyond odds ratios--communicating disease risk based on genetic profiles. Nat. Rev. Genet. 10, 264–269 (2009).
Weiss, S.T. et al. Creating and evaluating genetic tests predictive of drug response. Nat. Rev. Drug Discov. 7, 568–574 (2008).
Selzner, N. & McGilvray, I. Can genetic variations predict HCV treatment outcomes? J. Hepatol. 49, 494–497 (2008).
Aurora, R., Donlin, M.J., Cannon, N.A. & Tavis, J.E. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J. Clin. Invest. 119, 225–236 (2009).
Samarajiwa, S.A., Forster, S., Auchettl, K. & Hertzog, P.J. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37, D852–D857 (2009).
de Bakker, P.I.W. et al. Efficiency and power in genetic association studies. Nat. Genet. 37, 1217–1223 (2005).
Li, M., Liu, X., Zhou, Y. & Su, S.B. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J. Leukoc. Biol. 86, 23–32 (2009).
Mihm, S. et al. Interferon type I gene expression in chronic hepatitis C. Lab. Invest. 84, 1148–1159 (2004).
Fox, B.A., Sheppard, P.O. & O'Hara, P.J. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS One 4, e4933 (2009).
Coccia, E.M. et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34, 796–805 (2004).
Ank, N. et al. Lambda interferon (IFNλ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80, 4501–4509 (2006).
Doyle, S.E. et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 44, 896–906 (2006).
Sommereyns, C. et al. IFN-lambda is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4, e1000017 (2008).
Ank, N. et al. An Important Role for Type III Interferon (IFNλ/IL-28) in TLR-Induced Antiviral Activity. J. Immunol. 180, 2474–2485 (2008).
Marcello, T. et al. Interferons a and l inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 131, 1887–1898 (2006).
Sirén, J. et al. IFN-a regulates TLR-dependent gene expression of IFN-a, IFN-b, IL-28 and IL-29. J. Immunol. 174, 1932–1937 (2005).
Robek, M.D. et al. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79, 3851–3854 (2005).
Sheppard, P. et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4, 63–68 (2003).
Österlund, P.I. et al. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 179, 3434–3442 (2007).
Dellgren, C. et al. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 10, 125–131 (2009).
Zhu, H. & Liu, C. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J. Virol. 77, 5493–5498 (2003).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Saito, A. & Kamatani, N. Strategies for genome-wide association studies: optimization of study designs by the stepwise focusing method. J. Hum. Genet. 47, 360–365 (2002).
Skol, A.D. et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
Price, A.L. et al. Principal component analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Patterson, N.J., Price, A.L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Bersaglieri, T. et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 (2004).
Coelho, M. et al. Microsatellite variation and evolution of human lactase persistence. Hum. Genet. 117, 329–339 (2005).
WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Tian, C. et al. Analysis and application of European genetic substructure using 300K SNP information. PLoS Genet. 4, e4 (2008).
Sasieni, P.D. From genotype to genes: doubling the sample size. Biometrics 53, 1253–1261 (1997).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Parcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Gen. 81, 559–575 (2007).
Acknowledgements
V.S., D.R.B., G.J.S. and J.G. were supported by National Health and Medical Research Council Project grant 402577 and the Robert W. Storr Bequest to the University of Sydney. M.Bahlo has a National Health and Medical Research Council Career Development Award fellowship. T.B. is supported by the German Competence Network for Viral Hepatitis (Hep-Net), funded by the German Ministry of Education and Research (BMBF, grant number 01 KI 0437), the EU-Vigilanz network of excellence combating viral resistance (VIRGIL, project number LSHM-CT-2004-503359), and the BMBF project Host and viral determinants for susceptibility and resistance to hepatitis C virus infection (FKZ: 01KI0411; project B). D.S. and M.Bassendine are funded by a Medical Research Council UK project grant G0502028. We would like to thank all subjects for their valuable participation in this study.
Author information
Authors and Affiliations
Consortia
Contributions
V.S., G.J.S., D.R.B. and J.G. designed the study and wrote the manuscript. V.S. did the genotyping. M.M., M.Bahlo, and V.S. did the statistical analysis. Samples were phenotyped and collected by J.G., T.M., V.F., A.S., D.S., S.R., E.P., G.J.D., U.S., M.Bassendine., M.L.A., M.W., T.B. and G.A. All authors read and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have filed a provisional patent application on the intention of designing a genetic test kit based on the variants described in this paper.
Additional information
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4, Supplementary Figures 1 and 2 and Supplementary Note (PDF 2378 kb)
Rights and permissions
About this article
Cite this article
Suppiah, V., Moldovan, M., Ahlenstiel, G. et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet 41, 1100–1104 (2009). https://doi.org/10.1038/ng.447
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.447
This article is cited by
-
Response to antiviral therapy for chronic hepatitis C and risk of hepatocellular carcinoma occurrence in Japan: a systematic review and meta-analysis of observational studies
Scientific Reports (2023)
-
The presence of interferon affects the progression of non-alcoholic fatty liver disease
Genes & Immunity (2022)
-
The role of IFNL4 in liver inflammation and progression of fibrosis
Genes & Immunity (2022)
-
Recipient IL28B genotype CT is a predictor of new onset diabetes mellitus in liver transplant patients with chronic hepatitis C
International Journal of Diabetes in Developing Countries (2022)
-
Polymorphism rs368234815 of interferon lambda 4 gene and spontaneous clearance of hepatitis C virus in haemodialysis patients: a case-control study
BMC Infectious Diseases (2021)