Abstract
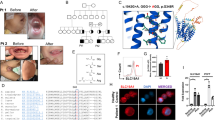
Patients with a combined immunodeficiency characterized by normal numbers but impaired function of T and B cells had a homozygous p.Tyr20His substitution in transferrin receptor 1 (TfR1), encoded by TFRC. The substitution disrupts the TfR1 internalization motif, resulting in defective receptor endocytosis and markedly increased TfR1 expression on the cell surface. Iron citrate rescued the lymphocyte defects, and expression of wild-type but not mutant TfR1 rescued impaired transferrin uptake in patient-derived fibroblasts. TfrcY20H/Y20H mice recapitulated the immunological defects of patients. Despite the critical role of TfR1 in erythrocyte development and function, patients had only mild anemia and only slightly increased TfR1 expression in erythroid precursors. We show that STEAP3, a metalloreductase expressed in erythroblasts, associates with TfR1 and partially rescues transferrin uptake in patient-derived fibroblasts, suggesting that STEAP3 may provide an accessory TfR1 endocytosis signal that spares patients from severe anemia. These findings demonstrate the importance of TfR1 in adaptive immunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Al-Herz, W. et al. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front. Immunol. 5, 162 (2014).
Jabara, H.H., Fu, S.M., Geha, R.S. & Vercelli, D. CD40 and IgE: synergism between anti-CD40 monoclonal antibody and interleukin 4 in the induction of IgE synthesis by highly purified human B cells. J. Exp. Med. 172, 1861–1864 (1990).
McHeyzer-Williams, L.J. & McHeyzer-Williams, M.G. Antigen-specific memory B cell development. Annu. Rev. Immunol. 23, 487–513 (2005).
Jing, S.Q., Spencer, T., Miller, K., Hopkins, C. & Trowbridge, I.S. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J. Cell Biol. 110, 283–294 (1990).
Andrews, N.C. Iron homeostasis: insights from genetics and animal models. Nat. Rev. Genet. 1, 208–217 (2000).
McGraw, T.E. & Maxfield, F.R. Human transferrin receptor internalization is partially dependent upon an aromatic amino acid on the cytoplasmic domain. Cell Regul. 1, 369–377 (1990).
Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–414 (2006).
Neckers, L.M. & Cossman, J. Transferrin receptor induction in mitogen-stimulated human T lymphocytes is required for DNA synthesis and cell division and is regulated by interleukin 2. Proc. Natl. Acad. Sci. USA 80, 3494–3498 (1983).
Ned, R.M., Swat, W. & Andrews, N.C. Transferrin receptor 1 is differentially required in lymphocyte development. Blood 102, 3711–3718 (2003).
Arezes, J. et al. Non-transferrin-bound iron (NTBI) uptake by T lymphocytes: evidence for the selective acquisition of oligomeric ferric citrate species. PLoS One 8, e79870 (2013).
Pinto, J.P. et al. Physiological implications of NTBI uptake by T lymphocytes. Front. Pharmacol. 5, 24 (2014).
Levy, J.E., Jin, O., Fujiwara, Y., Kuo, F. & Andrews, N.C. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat. Genet. 21, 396–399 (1999).
Ohgami, R.S., Campagna, D.R., McDonald, A. & Fleming, M.D. The Steap proteins are metalloreductases. Blood 108, 1388–1394 (2006).
Lambe, T. et al. Identification of a Steap3 endosomal targeting motif essential for normal iron metabolism. Blood 113, 1805–1808 (2009).
Collawn, J.F. et al. YTRF is the conserved internalization signal of the transferrin receptor, and a second YTRF signal at position 31–34 enhances endocytosis. J. Biol. Chem. 268, 21686–21692 (1993).
Ohgami, R.S. et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet. 37, 1264–1269 (2005).
Lespagnol, A. et al. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 15, 1723–1733 (2008).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Sobel, E., Sengul, H. & Weeks, D.E. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum. Hered. 52, 121–131 (2001).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Jabara, H.H. et al. Induction of germ-line and mature C epsilon transcripts in human B cells stimulated with rIL-4 and EBV. J. Immunol. 145, 3468–3473 (1990).
Jabara, H.H., Brodeur, S.R. & Geha, R.S. Glucocorticoids upregulate CD40 ligand expression and induce CD40L-dependent immunoglobulin isotype switching. J. Clin. Invest. 107, 371–378 (2001).
Crotzer, V.L., Mabardy, A.S., Weiss, A. & Brodsky, F.M. T cell receptor engagement leads to phosphorylation of clathrin heavy chain during receptor internalization. J. Exp. Med. 199, 981–991 (2004).
Roebroek, A.J., Gordts, P.L. & Reekmans, S. Knock-in approaches. Methods Mol. Biol. 693, 257–275 (2011).
Acknowledgements
We thank F. Alkuraya, H. Oettgen and T. Chatila for valuable discussions and the immunology laboratory staff at the Faculty of Medicine of Kuwait University for technical assistance. This work was supported by US National Institutes of Health (NIH) grants AI-076210, AI-007512 (R.S.G.), 1K08AI116979-01 (J.C.) and DK-089705 (N.C.A.), a grant from the Dubai Harvard Foundation for Medical Research (R.S.G.), the Perkins Fund (R.S.G.), the Howard Hughes Medical Institute (L.M.K.), the Manton Center for Orphan Disease Research (L.M.K.), the Kuwait Foundation for Advancement of Sciences 2010-1302-05 (W.A.-H.), a Jeffrey Modell Foundation Translational Research Program Grant award (J.C.) and a Manton Center Pilot Award (J.C.). Microarray genotyping and Sanger DNA sequencing were performed in the Molecular Genetics Core Facility at Boston Children's Hospital, supported by US NIH grant P30-HD18655 through the Intellectual and Developmental Disabilities Research Center and US NIH grant P50-NS40828 through the Neuromuscular Disease Project. We acknowledge the US NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of the mouse CD1d tetramers.
Author information
Authors and Affiliations
Contributions
H.H.J. performed functional experiments on the index family and on the TfrcY20H/Y20H mouse model. S.E.B. identified the TFRC mutation in the index family and performed genetic experiments and genome-wide linkage and whole-genome sequencing analyses. J.C. generated and analyzed the TfrcY20H/Y20H mouse model together with W.B. and D.F., performed functional experiments on patient B1 and provided clinical care to the patients in the index family. N.R., M.J.M., H.B., C.S. and Z.-J.L. performed functional experiments on the index family. F.R. performed genetic experiments. S.H.A. and B.K.A.-R. provided ancestry-matched control DNA samples. H.A.-D., R.A., M.A.-S., A.V., E.S. and S.A. identified and provided clinical care for patient B1. E.G.D. provided tissue specimens from the affected patients in family A who had undergone HSCT. T.K.O. performed bioinformatics analysis. M.S.-V., N.C.A., L.D.N., M.D.F. and W.A.-H. gave critical advice. W.A.-H. ascertained and provided clinical care to the index family. H.H.J., S.E.B., J.C., L.M.K. and R.S.G. wrote the manuscript. R.S.G. and L.M.K. designed and coordinated the investigations. The final version of the manuscript was approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
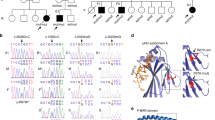
Supplementary Figure 1 Pedigrees of families A and B.
All numbered individuals were sequenced for the TFRC:c.58T>C mutation. Black-filled symbols indicate patients. Black dots indicate unaffected heterozygous carriers. Numbered subjects with open symbols lack the mutation. Subjects A1–A3 and B1 were studied in detail. With the exception of subjects A–G (for whom DNA was not available), A2 and A3 (for whom DNA was collected later), labeled subjects in families A and B were genotyped on the 6.0 Array for linkage analysis and/or haplotype analysis. The parents of subject B1 are first cousins once removed. SCT, stem cell transplantation.
Supplementary Figure 2 Evolutionary conservation of the TfR1 Tyr20 amino acid.
Protein sequence orthologous to human TfR1 was aligned in 81 non-human vertebrate species for which sequence was available, using Multiz alignment in the UCSC Genome Browser. The Tyr20 amino acid (arrow) is perfectly conserved, and the entire YTRF internalization motif is nearly perfectly conserved.
Supplementary Figure 3 Defective TfR1 internalization in patient cells.
(a) Representative FACS analysis and TfR1 surface expression shown by mean fluorescence intensity (MFI) on fibroblasts from patients A1–A3 compared to controls. (b) TfR1 internalization after cross-linking TfR1 using an anti-TfR1 monoclonal antibody and FITC-labeled secondary antibody on PHA-stimulated T cells, representing means ± SE from two independent experiments performed on separate blood draws from patient B1 and a control.
Supplementary Figure 4 TfR1 expression and in vitro T and B cell function following HSCT in patients A1–A3.
(a) TfR1 expression on T and B cells after HSCT from one experiment. (b) T cell proliferation in response to stimulation with anti-CD3 antibody and PMA + ionomycin following HSCT. (c) B cell proliferation and IgE production in response to stimulation with anti-CD40 antibody + IL-4 following HSCT. Data in b and c represent means ± SE of the three patients (Pt) and three controls (C) plated in duplicate, expressed as percent of controls. Patient B1 has not yet undergone HSCT.
Supplementary Figure 5 Generation of TfrcY20H/Y20H knock-in mice.
(a) Schematic of the wild-type Tfrc allele, the targeting construct and the targeted allele that has undergone homologous recombination in C57BL/6N embryonic stem cells. Tfrc exons are represented by blue boxes. The neomycin resistance gene (neo) under the control of the phosphoglycerate kinase 1 promoter was used as a positive selection marker; a diphtheria toxin A (DTA) fragment gene was used for negative selection. Red triangles, loxP sites. (b) Sanger sequencing demonstrating the homozygous c.58T>C mutation in DNA isolated from the tails of TfrcY20H/Y20H but not wild-type mice. (c) FACS analysis of TfR1 expression on CD3+ T cells and B220+ B cells from wild-type and TfrcY20H/Y20H mice.
Supplementary Figure 6 TfR1 surface expression on patient erythroid precursor cells.
(a) FACS analysis of TfR1 surface expression on glycophorin A (CD235a)+ erythroid precursor cells (EPCs). (b) Histograms for early (R1) and intermediate (R2) normoblasts.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–6 and Supplementary Note. (PDF 1057 kb)
Rights and permissions
About this article
Cite this article
Jabara, H., Boyden, S., Chou, J. et al. A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat Genet 48, 74–78 (2016). https://doi.org/10.1038/ng.3465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3465
This article is cited by
-
Mechanisms controlling cellular and systemic iron homeostasis
Nature Reviews Molecular Cell Biology (2024)
-
A Novel Homozygous Germline Mutation in Transferrin Receptor 1 (TfR1) Leads to Combined Immunodeficiency and Provides New Insights into Iron-Immunity Axis
Journal of Clinical Immunology (2024)
-
Impact of iron status on kidney outcomes in kidney transplant recipients
Scientific Reports (2023)
-
Nutrition impact on ILC3 maintenance and function centers on a cell-intrinsic CD71–iron axis
Nature Immunology (2023)
-
Ironman training for NK cells
Nature Immunology (2023)