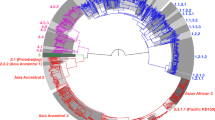
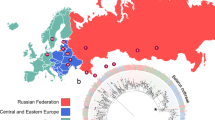
Abstract
Mycobacterium tuberculosis strains of the Beijing lineage are globally distributed and are associated with the massive spread of multidrug-resistant (MDR) tuberculosis in Eurasia. Here we reconstructed the biogeographical structure and evolutionary history of this lineage by genetic analysis of 4,987 isolates from 99 countries and whole-genome sequencing of 110 representative isolates. We show that this lineage initially originated in the Far East, from where it radiated worldwide in several waves. We detected successive increases in population size for this pathogen over the last 200 years, practically coinciding with the Industrial Revolution, the First World War and HIV epidemics. Two MDR clones of this lineage started to spread throughout central Asia and Russia concomitantly with the collapse of the public health system in the former Soviet Union. Mutations identified in genes putatively under positive selection and associated with virulence might have favored the expansion of the most successful branches of the lineage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Referenced accessions
NCBI Reference Sequence
Change history
10 February 2015
In the version of this article initially published online, affiliation 3 was incomplete, and the middle initials of author Michael Blum were inadvertently omitted. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Global Tuberculosis Report. (World Health Organization, Geneva, 2013).
Klopper, M. et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 19, 449–455 (2013).
Casali, N. et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46, 279–286 (2014).
Stoffels, K. et al. From multidrug- to extensively drug-resistant tuberculosis: upward trends as seen from a 15-year nationwide study. PLoS ONE 8, e63128 (2013).
Niemann, S. et al. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 48, 3544–3550 (2010).
Mokrousov, I. Insights into the origin, emergence, and current spread of a successful Russian clone of Mycobacterium tuberculosis. Clin. Microbiol. Rev. 26, 342–360 (2013).
Munsiff, S.S. et al. Persistence of a highly resistant strain of tuberculosis in New York City during 1990–1999. J. Infect. Dis. 188, 356–363 (2003).
Cowley, D. et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin. Infect. Dis. 47, 1252–1259 (2008).
Caminero, J.A. et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164, 1165–1170 (2001).
Ford, C.B. et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013).
Comas, I. & Gagneux, S. A role for systems epidemiology in tuberculosis research. Trends Microbiol. 19, 492–500 (2011).
Casali, N. et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22, 735–745 (2012).
Glynn, J.R., Whiteley, J., Bifani, P.J., Kremer, K. & van Soolingen, D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8, 843–849 (2002).
Bifani, P.J., Mathema, B., Kurepina, N.E. & Kreiswirth, B.N. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10, 45–52 (2002).
Parwati, I., van Crevel, R. & van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 (2010).
Hanekom, M. et al. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb.) 91, 510–523 (2011).
de Jong, B.C. et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 198, 1037–1043 (2008).
Kato-Maeda, M. et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin. Vaccine Immunol. 19, 1227–1237 (2012).
Weniger, T., Krawczyk, J., Supply, P., Niemann, S. & Harmsen, D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38, W326–W331 (2010).
Plikaytis, B.B. et al. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J. Clin. Microbiol. 32, 1542–1546 (1994).
Kalinowski, S.T. Counting alleles with rarefaction: private alleles and hierarchicalsampling design. Conserv. Genet. 5, 539–543 (2004).
Supply, P., Niemann, S. & Wirth, T. On the mutation rates of spoligotypes and variable numbers of tandem repeat loci of Mycobacterium tuberculosis. Infect. Genet. Evol. 11, 251–252 (2011).
Reyes, J.F. & Tanaka, M.M. Mutation rates of spoligotypes and variable numbers of tandem repeat loci in Mycobacterium tuberculosis. Infect. Genet. Evol. 10, 1046–1051 (2010).
Ragheb, M.N. et al. The mutation rate of mycobacterial repetitive unit loci in strains of M. tuberculosis from cynomolgus macaque infection. BMC Genomics 14, 145 (2013).
Comas, I., Homolka, S., Niemann, S. & Gagneux, S. Genotyping of genetically monomorphic bacteria: DNA sequencing in mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE 4, e7815 (2009).
Walker, T.M. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146 (2013).
Nieselt-Struwe, K. & von Haeseler, A. Quartet-mapping, a generalization of the likelihood-mapping procedure. Mol. Biol. Evol. 18, 1204–1219 (2001).
Namouchi, A., Didelot, X., Schock, U., Gicquel, B. & Rocha, E.P. After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22, 721–734 (2012).
Roetzer, A. et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 10, e1001387 (2013).
Ford, C.B. et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 43, 482–486 (2011).
Comas, I. & Gagneux, S. The past and future of tuberculosis research. PLoS Pathog. 5, e1000600 (2009).
Colditz, G.A. et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. J. Am. Med. Assoc. 271, 698–702 (1994).
Holden, C. Stalking a killer in Russia's prisons. Science 286, 1670 (1999).
Bifani, P.J. et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. J. Am. Med. Assoc. 275, 452–457 (1996).
Parish, T. et al. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140 (2003).
Duforet-Frebourg, N., Bazin, E. & Blum, M.G. Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol. Biol. Evol. 31, 2483–2495 (2014).
Maloney, E. et al. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5, e1000534 (2009).
Sirakova, T.D., Fitzmaurice, A.M. & Kolattukudy, P. Regulation of expression of mas and fadD28, two genes involved in production of dimycocerosyl phthiocerol, a virulence factor of Mycobacterium tuberculosis. J. Bacteriol. 184, 6796–6802 (2002).
Ebrahimi-Rad, M. et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9, 838–845 (2003).
Etienne, G. et al. Identification of the polyketide synthase involved in the biosynthesis of the surface-exposed lipooligosaccharides in mycobacteria. J. Bacteriol. 191, 2613–2621 (2009).
Ahmad, S., El-Shazly, S., Mustafa, A.S. & Al-Attiyah, R. Mammalian cell-entry proteins encoded by the mce3 operon of Mycobacterium tuberculosis are expressed during natural infection in humans. Scand. J. Immunol. 60, 382–391 (2004).
Li, X.Z., Zhang, L. & Nikaido, H. Efflux pump–mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423 (2004).
Meikle, V. et al. Identification of novel Mycobacterium bovis antigens by dissection of crude protein fractions. Clin. Vaccine Immunol. 16, 1352–1359 (2009).
de Vos, M. et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob. Agents Chemother. 57, 827–832 (2013).
Comas, I. et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110 (2012).
Comas, I. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013).
Fuller, D.Q. et al. The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 323, 1607–1610 (2009).
Fang, J. Atlas for Sustainability in Polynesian Island Cultures and Ecosystems (Sea Education Association, 2013).
Laruelle, M. & Peyrouse, S. Cross-border minorities as cultural and economic mediators between China and Central Asia. China and Eurasia Forum Quarterly 7, 93–119 (2009).
Drolet, G.J. World War I and tuberculosis. A statistical summary and review. Am. J. Public Health Nations Health 35, 689–697 (1945).
Bryant, J.M. et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect. Dis. 13, 110 (2013).
Aguilar, D. et al. Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb.) 90, 319–325 (2010).
Ribeiro, S.C. et al. Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J. Clin. Microbiol. 52, 2615–2624 (2014).
Gioffré, A. et al. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 7, 325–334 (2005).
Stavrum, R. et al. Modulation of transcriptional and inflammatory responses in murine macrophages by the Mycobacterium tuberculosis mammalian cell entry (Mce) 1 complex. PLoS ONE 6, e26295 (2011).
Ahidjo, B.A. et al. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE 6, e21738 (2011).
Osório, N.S. et al. Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic- and nonantibiotic-related pressure. Mol. Biol. Evol. 30, 1326–1336 (2013).
Zhang, H. et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45, 1255–1260 (2013).
Farhat, M.R. et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189 (2013).
Supply, P. et al. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39, 3563–3571 (2001).
Supply, P. et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit–variable number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 4498–4510 (2006).
Kalinowski, S.T. HP-rare: a computer program for performing rarefaction on measures of allelic diversity. Mol. Ecol. Notes 5, 187–189 (2005).
Beaumont, M.A. Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029 (1999).
Wilson, I.J., Weale, M.E. & Balding, D.J. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. Ser. A Stat. Soc. 166, 155–188 (2003).
Wilson, I.J. & Balding, D.J. Genealogical inference from microsatellite data. Genetics 150, 499–510 (1998).
Ohta, T. & Kimura, M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 22, 201–204 (1973).
Metropolis, N., Rosenbluth, A.W., Rosenbluth, M.N., Teller, A.H. & Teller, E. Equations of state calculations by fast computing machine. J. Chem. Phys. 21, 1087–1091 (1953).
Hastings, W.K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970).
Wirth, T. et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4, e1000160 (2008).
Blom, J. et al. Exact and complete short-read alignment to microbial genomes using Graphics Processing Unit programming. Bioinformatics 27, 1351–1358 (2011).
Schmidt, H.A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Posada, D. & Crandall, K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998).
Hasegawa, M., Kishino, H. & Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985).
Drummond, A.J., Rambaut, A., Shapiro, B. & Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005).
Drummond, A.J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Casali, N. & Riley, L.W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60 (2007).
Zhang, Z. et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263 (2006).
Acknowledgements
We gratefully acknowledge L. Cowan and J. Posey (US Centers for Disease Control and Prevention) for providing us with significant amounts of genotyping data for M. tuberculosis Beijing isolates. We thank T. Ubben, I. Radzio, T. Struwe-Sonnenschein and J. Zallet (Research Center Borstel) for excellent technical assistance. We acknowledge J. Peh for her assistance and support in the study and I. Comas for statistical advice. Parts of this work have been supported by grants from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the European Union PathoNGenTrace project and grant agreement 223681 in the framework of the TB-PAN-NET project. We also thank Action Transversale du Muséum National d'Histoire Naturelle 'Les Microorganismes, Acteurs Clés dans les Ecosystèmes' for financial support. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
I.M., P. Supply, S.N. and T.W. designed the study. M.M., P. Supply, S.N. and T.W. analyzed data and wrote the manuscript with comments from all authors. M.M., C.B., S. Mona and T.W. performed population genetics and phylogenetic analyses. M.M., N.D.-F., M.G.B.B. and T.W. conducted selection tests. T.A.K. performed whole-genome sequencing and SNP calling. P. Supply, M.M., E.W., S.L., S.R.-G., I.M., S.N., E.A., C.A.-B., A.A., E.A.-K., M. Ballif, F.B., H.P.B., C.E.B., M. Bonnet, E.B., I.C.-H., D.C., H.C., S.C., V.C., R.D., F.D., M.F.-D., S. Gagneux, S. Ghebremichael, M.H., S.H., W.-w.J., S.K., I.K., T.L., S. Maeda, V.N., M.R., N.R., S.S., E.S.-P., B.S., I.C.S., A.S., L.-H.S., P. Stakenas, K.T., F.V., D.V., C.W. and R.W. obtained mycobacterial genotyping data and drug susceptibility test results.
Corresponding authors
Ethics declarations
Competing interests
P. Supply is a consultant for Genoscreen. C.A.-B. and C.W. were or are employees of the same company. The other authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 2–10 and 12. (PDF 15198 kb)
Supplementary Table 1
Genotyping results (24-locus MIRU-VNTR), country of origin, geolocalization and available phenotypic drug susceptibility test results for 4,987 analyzed clinical isolates from the MTBC Beijing lineage. (XLSX 897 kb)
Supplementary Table 11
Top Bayes factor variants. SNPs are given relative to the H37Rv reference sequence, and putative targets of selection are highlighted. The color codes along the logBF column correspond, respectively, to SNPs specific to the central Asia outbreak (blue), the European-Russian W148 outbreak (green) and the members of the modern Beijing lineage (orange). (XLSX 2913 kb)
Rights and permissions
About this article
Cite this article
Merker, M., Blin, C., Mona, S. et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47, 242–249 (2015). https://doi.org/10.1038/ng.3195
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3195
This article is cited by
-
Detection of a historic reservoir of bedaquiline/clofazimine resistance-associated variants in Mycobacterium tuberculosis
Genome Medicine (2024)
-
Genotypic and phenotypic comparison of drug resistance profiles of clinical multidrug-resistant Mycobacterium tuberculosis isolates using whole genome sequencing in Latvia
BMC Infectious Diseases (2023)
-
Strain structure analysis of Mycobacterium tuberculosis circulating among HIV negative, positive and drug resistant TB patients attending chest clinics in Western Kenya
BMC Pulmonary Medicine (2023)
-
Will the Russian war in Ukraine unleash larger epidemics of HIV, TB and associated conditions and diseases in Ukraine?
Harm Reduction Journal (2023)
-
Genotyping of Mycobacterium tuberculosis complex isolated from humans and animals in northeastern Iran
Scientific Reports (2023)