Abstract
Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) is a rare, highly aggressive form of ovarian cancer primarily diagnosed in young women. We identified inactivating biallelic SMARCA4 mutations in 100% of the 12 SCCOHT tumors examined. Protein studies confirmed loss of SMARCA4 expression, suggesting a key role for the SWI/SNF chromatin-remodeling complex in SCCOHT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Estel, R., Hackethal, A., Kalder, M. & Munstedt, K. Arch. Gynecol. Obstet. 284, 1277–1282 (2011).
Young, R.H., Oliva, E. & Scully, R.E. Am. J. Surg. Pathol. 18, 1102–1116 (1994).
Harrison, M.L. et al. Gynecol. Oncol. 100, 233–238 (2006).
Seidman, J.D. Gynecol. Oncol. 59, 283–287 (1995).
Pautier, P. et al. Ann. Oncol. 18, 1985–1989 (2007).
McCluggage, W.G. Adv. Anat. Pathol. 11, 288–296 (2004).
Hendricks, K.B., Shanahan, F. & Lees, E. Mol. Cell. Biol. 24, 362–376 (2004).
Napolitano, M.A. et al. J. Cell Sci. 120, 2904–2911 (2007).
Kupryjańczyk, J. et al. Pol. J. Pathol. 64, 238–246 (2013).
Longy, M., Toulouse, C., Mage, P., Chauvergne, J. & Trojani, M. J. Med. Genet. 33, 333–335 (1996).
McDonald, J.M. et al. J. Pediatr. Surg. 47, 588–592 (2012).
Reisman, D., Glaros, S. & Thompson, E.A. Oncogene 28, 1653–1668 (2009).
Wilson, B.G. & Roberts, C.W. Nat. Rev. Cancer 11, 481–492 (2011).
Oike, T. et al. Cancer Res. 73, 5508–5518 (2013).
Guan, B., Wang, T.L. & Shih, I.-M. Cancer Res. 71, 6718–6727 (2011).
Won, H.H., Scott, S.N., Brannon, A.R., Shah, R.H. & Berger, M.F. J. Vis. Exp. 80, e50710 (2013).
Wagle, N. et al. Cancer Discov. 2, 82–93 (2012).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. Nat. Genet. 43, 491–498 (2011).
Cibulskis, K. et al. Nat. Biotechnol. 31, 213–219 (2013).
Robinson, J.T. et al. Nat. Biotechnol. 29, 24–26 (2011).
Cerami, E. et al. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Sci. Signal. 6, pl1 (2013).
Acknowledgements
We appreciate the assistance of the Small Cell Ovarian Cancer Foundation with recruitment. We are grateful to all the patients and family members who generously donated biospecimens so that we could conduct this study. We thank G. Monemvasitis for editorial assistance. This work was supported by the Katie Oppo Research Fund, the Chia Family Foundation, the Geoffrey Beene Cancer Research Center, the Farmer Family Foundation and a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209).
Author information
Authors and Affiliations
Contributions
J.J.M. and D.A.L. conceived the study. N.S., M.F.B. and D.A.L. supervised analyses. P.J., J.J.M. and F.D. designed and performed analyses. R.A.S. and D.A.L. provided patient materials. N.O. processed tissue samples. R.A.S. performed pathology review. M.G. performed statistical analyses. S.N.S. and R.S. performed massively parallel DNA sequencing. J.G. performed computational analyses. P.J., J.J.M. and D.A.L. wrote the manuscript with contributions from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
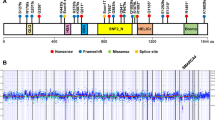
Supplementary Figure 1 Sequence analyses for SMARCA4 in SCCOHT cases.
Next-generation sequence coverage demonstrating identified variants (top panels) and validation through Sanger sequencing (bottom panels).
Supplementary Figure 2 SMARCA4 gene expression across TCGA tumors for cases with available mutation and RNA-seq data (RSEM).
A correlation is seen between inactivating SMARCA4 mutations and decreased gene expression across various solid tumors. A two-sided Student's t test was used to compare samples with non-missense mutations and other samples without mutations or with only missense mutations. For all TCGA samples, the mean RNA-seq RSEM (2,050, s.d. of 1,760) was less in samples with non-missense mutations than in other samples without mutations or with only missense mutations (3,724, s.d. of 1,692; P = 8.7 × 10−4). For TCGA lung adenocarcinoma samples, the mean RNA-seq RSEM (601, s.d. of 370) was less in samples with non-missense mutations than in other samples without mutations or with only missense mutations (3,330, s.d. of 1,524; P = 2 × 10−8).
Supplementary Figure 3 Immunohistochemistry for SMARCA4 in SCCOHT cases.
High-grade serous ovarian carcinoma is used as a positive control. Case numbers are indicated in each panel. Immunohistochemistry results are provided in Supplementary Table 1. Note the intense staining of blood vessels and stromal cell nuclei as internal controls.
Supplementary Figure 4 Analysis of homozygous deletion in case 103.
Next-generation sequence coverage demonstrating that exons 25 and 26 are deleted. An electropherogram from Sanger sequencing of cDNA validating that the deletion retains an ORF from exon 24 to exon 27 (Panel A). One-step RT-PCR confirms that tumor tissue yields a single band with primers that span exons 24 and 27 (Panel B; *, nonspecific band). One-step RT-PCR with primers targeting regions upstream and downstream from the deletion site show equal expression, demonstrating continuation of transcription downstream from the deletion (Panel C).
Supplementary Figure 5 Analysis of splice-site variant in case 102.
One-step RT-PCR confirms that the exon-intron band is preferentially expressed over the exon-exon band in tumor tissue (Panel A). One-step RT-PCR with primers targeting regions upstream and downstream from the mutation site show equal expression, demonstrating continuation of transcription downstream from the mutation (Panel B). Immunoblots are shown in Figure 2b. The exon-exon primers detected weaker bands, reflecting loss of expression in tumor tissues compared with normal tissues in cases with splice-site mutations. The exon-intron primers demonstrated equivalent to greater expression of the retained intron in the tumor tissues. As SMARCA4 introns may be retained in non-cancer tissues, some intronic expression is expected in normal tissues. These data taken together indicate preferential intronic expression, as expected, in cDNA sequenced from tumor samples with splice-site mutations.
Supplementary Figure 6 Sequence analyses for SMARCA4 in the H1299 cell line.
An electropherogram from Sanger sequencing of genomic DNA validating a 69-nt deletion in the ORF of this control cell line that results in loss of protein expression, as shown in Figure 2b.
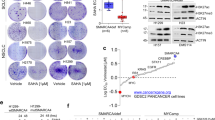
Supplementary Figure 7 SMARCA4 effect on cell proliferation.
SMARCA4 overexpression in H1299 cells. Representative immunoblot from three biologic replicates demonstrates a correlation between increased SMARCA4 and p21 expression (Panel A). Cell growth assessment in H1299 cells overexpressing SMARCA4. Mean cell counts from three biologic replicates (Panel B). Representative immunoblot confirmed SMARCA4 knockdown in 293T cells using shRNA. As a control, shNTC (Non-Targeting Control) was used (Panel C). XTT proliferation assay in 293T cells depleted of SMARCA4. Means represent three independent experiments (Panel D).
Supplementary Figure 8 Overall survival among lung adenocarcinoma TCGA cases based on inactivating SMARCA4 mutations.
Median overall survival was 11.6 months among 6 patients with inactivating SMARCA4 mutations compared with 44.6 months for 197 patients without inactivating mutations.
Supplementary Figure 9 Histopathological features of an SCCOHT.
The typical histopathological features of SCCOHT, including a combination of small neoplastic cells forming a pseudofollicular space and larger rhabdoid cells, are visible in a sample obtained from 1 of 12 tumors that were subjected to target capture and massively parallel DNA sequencing (hematoxylin and eosin).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–5 (PDF 5232 kb)
Source data
Rights and permissions
About this article
Cite this article
Jelinic, P., Mueller, J., Olvera, N. et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet 46, 424–426 (2014). https://doi.org/10.1038/ng.2922
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2922
This article is cited by
-
Alanine supplementation exploits glutamine dependency induced by SMARCA4/2-loss
Nature Communications (2023)
-
Primary cutaneous SMARCA4-deficient undifferentiated malignant neoplasm: first two cases with clinicopathologic and molecular comparison to eight visceral counterparts
Modern Pathology (2022)
-
HELLS serves as a poor prognostic biomarker and its downregulation reserves the malignant phenotype in pancreatic cancer
BMC Medical Genomics (2021)
-
DICER1-associated sarcomas: towards a unified nomenclature
Modern Pathology (2021)
-
Genomic alterations in gynecological malignancies: histotype-associated driver mutations, molecular subtyping schemes, and tumorigenic mechanisms
Journal of Human Genetics (2021)