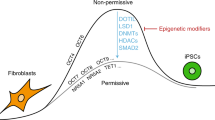
Abstract
Vitamin C, a micronutrient known for its anti-scurvy activity in humans, promotes the generation of induced pluripotent stem cells (iPSCs)1 through the activity of histone demethylating dioxygenases2,3. TET hydroxylases are also dioxygenases implicated in active DNA demethylation4,5,6,7,8. Here we report that TET1 either positively or negatively regulates somatic cell reprogramming depending on the absence or presence of vitamin C. TET1 deficiency enhances reprogramming, and its overexpression impairs reprogramming in the context of vitamin C2,9 by modulating the obligatory mesenchymal-to-epithelial transition (MET)10,11. In the absence of vitamin C, TET1 promotes somatic cell reprogramming independent of MET. Consistently, TET1 regulates 5-hydroxymethylcytosine (5hmC) formation at loci critical for MET in a vitamin C–dependent fashion. Our findings suggest that vitamin C has a vital role in determining the biological outcome of TET1 function at the cellular level. Given its benefit to human health, vitamin C should be investigated further for its role in epigenetic regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Esteban, M.A. et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79 (2010).
Chen, J. et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 (2013).
Wang, T. et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9, 575–587 (2011).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
He, Y.-F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011).
Chen, J. et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 21, 884–894 (2011).
Li, R. et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 (2010).
Samavarchi-Tehrani, P. et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010).
Zhang, R.R. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 (2013).
Nakagawa, M. et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 (2008).
Szabó, P.E., Hubner, K., Scholer, H. & Mann, J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115, 157–160 (2002).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Gillich, A. et al. Epiblast stem cell–based system reveals reprogramming synergy of germline factors. Cell Stem Cell 10, 425–439 (2012).
Sridharan, R. et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 (2009).
Silva, J. et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253 (2008).
Maherali, N. et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 (2007).
Gao, Y. et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 (2013).
Costa, Y. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013).
Wang, T. et al. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine–mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol. 15, 700–711 (2013).
Sotiriou, S. et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat. Med. 8, 514–517 (2002).
Stadtfeld, M. et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all–iPS cell mice from terminally differentiated B cells. Nat. Genet. 44, 398–405 (2012).
Doege, C.A. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 (2012).
Mao, Y. et al. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 138, 947–957 (2011).
Minor, E.A., Court, B.L., Young, J.I. & Wang, G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase–mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288, 13669–13674 (2013).
Yin, R. et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135, 10396–10403 (2013).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Dawlaty, M.M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Yildirim, O. et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510 (2011).
Gu, T.-P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Koh, K.P. et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213 (2011).
Monfort, A. & Wutz, A. Breathing-in epigenetic change with vitamin C. EMBO Rep. 14, 337–346 (2013).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Acknowledgements
We are grateful to J. Nie, J. Zhu, B. Wei, X. Li, T. Peng, X. Huang and C. Tan for technical assistance. We thank H. Zheng for constructive criticism. We deeply appreciate the work of X. Zhao, H. Song, S. Chu and Y. Chen in blastocyst injection, and we thank R. Luo for isolating MEFs. We thank S. Gao (National Institute of Biological Sciences, China) for providing the FUW-TetOn-Tet1 vector. We wish to thank all members of our laboratories for their support. This work is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA01020401), the National Basic Research Program of China (grants 2014CB965200, 2012CB966802 and 2011CB965204), Guangdong Natural Science Funds for Distinguished Young Scholars (grant S2013050014040), the National Natural Science Foundation of China (grants 31271357, 31230039, 31221001 and 91213304), the Ministry of Science and Technology International Technology Cooperation Program (grant 2012DFH30050), the Science and Technology Planning Project of Guangdong Province (grant 2011A060901019), the National Science & Technology Major Special Project on Major New Drug Innovation (grant 2011ZX09102-010), the Youth Innovation Promotion Association of the Chinese Academy of Sciences and the Guangzhou Pearl River Nova program (grant 2012J2200070).
Author information
Authors and Affiliations
Contributions
J.C., L.G. and L.Z. initiated the study, designed and performed experiments and analyzed data. Haoyu Wu performed the experiments and analyzed data. J.Y. and H.L. performed qPCR and chromatin immunoprecipitation experiments and prepared some cell samples. J.C. and X.W. performed bioinformatics analysis. X.H. performed reprogramming assays and chromatin immunoprecipitation. T.G. generated the antibody to TET1. Z.Z. bred the Tet1-null mice and derived primary cells. Jing Liu isolated primary cells and performed reprogramming assays. Jiadong Liu performed bisulfite genomic sequencing. Hongling Wu performed proliferation and apoptosis assays. S.-Q.M. generated the Tet1-null mice. K.M. performed blastocyst injections. Y.L. produced recombinant growth factor and assisted in antibody production. K.L. performed karyotype analysis. J.Q. performed protein blotting. H.Y. assisted in the construction of sequencing libraries. G.P. assisted in next-generation sequencing. G.-L.X. and D.P. conceived and supervised the entire study. D.P. and J.C. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 6 and 7 (PDF 6993 kb)
Supplementary Tables 1–5
Supplementary Tables 1–5 (XLSX 1895 kb)
Rights and permissions
About this article
Cite this article
Chen, J., Guo, L., Zhang, L. et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet 45, 1504–1509 (2013). https://doi.org/10.1038/ng.2807
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2807
This article is cited by
-
Obesity and dyslipidemia are associated with partially reversible modifications to DNA hydroxymethylation of apoptosis- and senescence-related genes in swine adipose-derived mesenchymal stem/stromal cells
Stem Cell Research & Therapy (2023)
-
Unraveling the 2,3-diketo-l-gulonic acid-dependent and -independent impacts of l-ascorbic acid on somatic cell reprogramming
Cell & Bioscience (2023)
-
Nuclear localization of TET2 requires β-catenin activation and correlates with favourable prognosis in colorectal cancer
Cell Death & Disease (2023)
-
Epigenetic reprogramming of cell identity: lessons from development for regenerative medicine
Clinical Epigenetics (2021)
-
Proteins in DNA methylation and their role in neural stem cell proliferation and differentiation
Cell Regeneration (2021)