Abstract
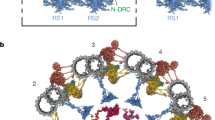
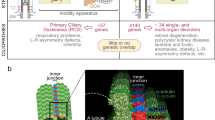
Primary ciliary dyskinesia (PCD) is characterized by dysfunction of respiratory cilia and sperm flagella and random determination of visceral asymmetry. Here, we identify the DRC1 subunit of the nexin-dynein regulatory complex (N-DRC), an axonemal structure critical for the regulation of dynein motors, and show that mutations in the gene encoding DRC1, CCDC164, are involved in PCD pathogenesis. Loss-of-function mutations disrupting DRC1 result in severe defects in assembly of the N-DRC structure and defective ciliary movement in Chlamydomonas reinhardtii and humans. Our results highlight a role for N-DRC integrity in regulating ciliary beating and provide the first direct evidence that mutations in DRC genes cause human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Referenced accessions
NCBI Reference Sequence
Change history
08 February 2013
In the version of this article initially published online, authors Unne Stenram and Birgitta Carlén were listed with the incorrect departmental affiliation. Their correct affiliation is the Department of Pathology, Lund University and Skane University Hospital, Lund, Sweden, not the Department of Otorhinolaryngology–Head and Neck Surgery, Lund University and Skane University Hospital, Lund, Sweden, as originally stated. The error has been corrected for the print, PDF and HTML versions of this article.
References
Fliegauf, M., Benzing, T. & Omran, H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 (2007).
Zariwala, M.A., Omran, H. & Ferkol, T.W. The emerging genetics of primary ciliary dyskinesia. Proc. Am. Thorac. Soc. 8, 430–433 (2011).
Mazor, M. et al. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 88, 599–607 (2011).
Olbrich, H. et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 30, 143–144 (2002).
Panizzi, J.R. et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 44, 714–719 (2012).
Smith, E.F. & Yang, P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton 57, 8–17 (2004).
Huang, B., Ramanis, Z. & Luck, D.J. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28, 115–124 (1982).
Piperno, G., Mead, K., LeDizet, M. & Moscatelli, A. Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 125, 1109–1117 (1994).
Piperno, G., Mead, K. & Shestak, W. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118, 1455–1463 (1992).
Becker-Heck, A. et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 43, 79–84 (2011).
Merveille, A.C. et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 43, 72–78 (2011).
O'Toole, E.T., Giddings, T.H. Jr., Porter, M.E. & Ostrowski, L.E. Computer-assisted image analysis of human cilia and Chlamydomonas flagella reveals both similarities and differences in axoneme structure. Cytoskeleton (Hoboken) 69, 577–590 (2012).
Heuser, T., Raytchev, M., Krell, J., Porter, M.E. & Nicastro, D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187, 921–933 (2009).
Gardner, L.C., O'Toole, E., Perrone, C.A., Giddings, T. & Porter, M.E. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell Biol. 127, 1311–1325 (1994).
Brokaw, C.J. & Kamiya, R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton 8, 68–75 (1987).
Kabututu, Z.P., Thayer, M., Melehani, J.H. & Hill, K.L. CMF70 is a subunit of the dynein regulatory complex. J. Cell Sci. 123, 3587–3595 (2010).
Rupp, G. & Porter, M.E. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest–specific gene product. J. Cell Biol. 162, 47–57 (2003).
Colantonio, J.R. et al. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature 457, 205–209 (2009).
Lin, J. et al. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286, 29175–29191 (2011).
Silflow, C.D. et al. Expression of flagellar protein genes during flagellar regeneration in Chlamydomonas. Cold Spring Harb. Symp. Quant. Biol. 46, 157–169 (1982).
Dymek, E.E., Heuser, T., Nicastro, D. & Smith, E.F. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol. Biol. Cell 22, 2520–2531 (2011).
Yanagisawa, H.A. & Kamiya, R. A tektin homologue is decreased in Chlamydomonas mutants lacking an axonemal inner-arm dynein. Mol. Biol. Cell 15, 2105–2115 (2004).
Yagi, T., Uematsu, K., Liu, Z. & Kamiya, R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J. Cell Sci. 122, 1306–1314 (2009).
Bui, K.H., Yagi, T., Yamamoto, R., Kamiya, R. & Ishikawa, T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 198, 913–925 (2012).
Zadro, C. et al. Five new OTOF gene mutations and auditory neuropathy. Int. J. Pediatr. Otorhinolaryngol. 74, 494–498 (2010).
Mateos-Corral, D., Coombs, R., Grasemann, H., Ratjen, F. & Dell, S.D. Diagnostic value of nasal nitric oxide measured with non-velum closure techniques for children with primary ciliary dyskinesia. J. Pediatr. 159, 420–424 (2011).
Carlén, B., Lindberg, S. & Stenram, U. Absence of nexin links as a possible cause of primary ciliary dyskinesia. Ultrastruct. Pathol. 27, 123–126 (2003).
Bui, K.H., Sakakibara, H., Movassagh, T., Oiwa, K. & Ishikawa, T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 183, 923–932 (2008).
Blanchon, S. et al. Delineation of CCDC39/CCDC40 mutation spectrum and associated phenotypes in primary ciliary dyskinesia. J. Med. Genet. 49, 410–416 (2012).
Fliegauf, M. et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 171, 1343–1349 (2005).
Pazour, G.J., Agrin, N., Walker, B.L. & Witman, G.B. Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J. Med. Genet. 43, 62–73 (2006).
Kathir, P. et al. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2, 362–379 (2003).
Nelson, J.A., Savereide, P.B. & Lefebvre, P.A. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14, 4011–4019 (1994).
Harris, E. Chlamydomonas in the laboratory. in The Chlamydomonas Sourcebook Vol. 1 (ed. Harris, E.) Ch. 8, 241–301 (Academic Press, San Diego, 2009).
Witman, G.B. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280–290 (1986).
Bower, R. et al. IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol. Biol. Cell 20, 3055–3063 (2009).
Barbato, A. et al. Primary ciliary dyskinesia: a consensus statement on diagnostic and treatment approaches in children. Eur. Respir. J. 34, 1264–1276 (2009).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Acknowledgements
We thank G. Piperno for providing the peptide sequences used to identify the Chlamydomonas DRC1 gene. M.E.P. also thanks C. Perrone, T. vy Le, A. Bostrom, J. Mueller and J.A. Knott for assistance with mapping the DRC1 locus, P. Kathir, C. Silflow and S. Dutcher for advice on RFLP mapping and T. Markowski and B. Witthun for assistance with mass spectrometry and spectral counting. We thank the German patient support group Kartagener Syndrom und Primaere Ciliaere Dyskinesie e.V. We also thank A. Heer and C. Westermann for excellent technical assistance. We thank M. Laudon and the Chlamydomonas Genetics Center for strains. For antibodies, we thank R. Linck (University of Minnesota) for antibody to Rib72, M. Sanders (University of Minnesota) for MC1 antibody to centrin, R. Kamiya (University of Tokyo) for antibodies to tektin, p44 and p38, T. Yagi (Kyoto University) for antibodies to DHC5, DHC9 and DHC11, P. Yang (Marquette University) for antibody to RSP16, E. Smith (Dartmouth College) for antibody to CaM-IP3 and G. Piperno (Mount Sinai School of Medicine) for antibody to p28). This work was supported by US National Institutes of Health (NIH) grants to M.E.P. (GM-55667) and W.S.S. (GM-051173), a National Research Service Award (NRSA) postdoctoral fellowship to M.W. (GM-075446), funding to W.S.S. from the Children's Healthcare of Atlanta and Emory University School of Medicine Pediatric Research Center and funding to H. Omran (Deutsche Forschungsgemeinschaft DFG Om 6/4 and Om 6/5, GRK1104, SFB592, IZKF Muenster and the Cell Dynamics and Disease (CEDAD) graduate school as well as SYSCILIA from the European Community). The Center for Mass Spectrometry and Proteomics at the University of Minnesota is supported by multiple grants, including National Science Foundation (NSF) Major Research Instrumentation grants 9871237 and NSF-DBI-0215759. Technical and software support was also provided by the Minnesota Supercomputing Institute.
Author information
Authors and Affiliations
Contributions
D.T. and M.W. cloned the Chlamydomonas DRC1 gene and generated the antibody against the DRC1 protein. D.T. identified the mutation in the pf3 strain. R.B. performed biochemical studies on Chlamydomonas axonemes. M.E.P. evaluated the spectral counting and pf3 mapping data. H. Olbrich evaluated linkage analysis and performed sequencing of human subjects with PCD. C.W. evaluated clinical data from individuals with PCD and performed high-speed video microscopy analysis and nasal NO measurements on OP-26II1. N.T.L. performed high-resolution immunofluorescence microscopy of PCD samples. P.P. generated in situ hybridization of Ccdc164 at the mouse embryonic node and performed immunofluorescence. H. Olbrich, N.T.L., P.P., D.T., R.B. and M.W. prepared the figures. S.L., U.S. and B.C. provided clinical data, TEM and DNA from OP-39 and OP-56. S.L. performed surgery on OP-59II1. E.H. provided clinical data and DNA from OP-26. G.K. performed TEM. P.N. and G.N. performed linkage and haplotype analyses. H. Omran evaluated all TEM analyses. H. Omran, W.S.S. and M.E.P. coordinated the study. M.W., M.E.P. and H. Omran wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–5 and Supplementary Note (PDF 1440 kb)
Supplementary Video 1
Respiratory cilia of individual OP-26II1 in real time (AVI 15106 kb)
Supplementary Video 2
Respiratory cilia of individual OP-26II1 in slow motion (1/8 speed) (WMV 1770 kb)
Supplementary Video 3
Respiratory cilia of a healthy control in real time (WMV 1120 kb)
Supplementary Video 4
Respiratory cilia of a healthy control in slow motion (1/8 speed) (WMV 4355 kb)
Rights and permissions
About this article
Cite this article
Wirschell, M., Olbrich, H., Werner, C. et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet 45, 262–268 (2013). https://doi.org/10.1038/ng.2533
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2533
This article is cited by
-
Prevalence and founder effect of DRC1 exon 1–4 deletion in Korean patients with primary ciliary dyskinesia
Journal of Human Genetics (2023)
-
Spokes and barrels tune the asymmetric beating of mammalian sperm flagella
Nature Structural & Molecular Biology (2023)
-
Lack of CFAP54 causes primary ciliary dyskinesia in a mouse model and human patients
Frontiers of Medicine (2023)
-
Characterization of a DRC1 null variant associated with primary ciliary dyskinesia and female infertility
Journal of Assisted Reproduction and Genetics (2023)
-
Bi-allelic variants in human TCTE1/DRC5 cause asthenospermia and male infertility
European Journal of Human Genetics (2022)