Abstract
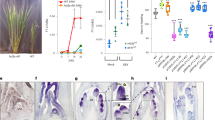
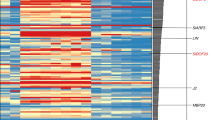
The transition to flowering is a major determinant of plant architecture, and variation in the timing of flowering can have profound effects on inflorescence architecture, flower production and yield. Here, we show that the tomato mutant terminating flower (tmf) flowers early and converts the multiflowered inflorescence into a solitary flower as a result of precocious activation of a conserved floral specification complex encoded by ANANTHA (AN) and FALSIFLORA (FA). Without TMF, the coordinated flowering process is disrupted, causing floral identity genes, such as AN and members of the SEPALLATA (SEP) family, to activate precociously, while the expression of flowering transition genes, such as FRUITFULL (FUL), is delayed. Indeed, driving AN expression precociously is sufficient to cause early flowering, and this expression transforms multiflowered inflorescences into normal solitary flowers resembling those of the Solanaceae species petunia and tobacco. Thus, by timing AN activation, TMF synchronizes flower formation with the gradual reproductive transition, which, in turn, has a key role in determining simple versus complex inflorescences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Weberling, F. Morphology of Flowers and Inflorescences (Cambridge University Press, Cambridge, 1989).
Kobayashi, Y. & Weigel, D. Move on up, it′s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384 (2007).
Turck, F., Fornara, F. & Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 (2008).
Bell, A.D. Plant Form: An Illustrated Guide to Flowering Plant Morphology (Oxford University Press, Oxford, 1993).
Knapp, S., Bohs, L., Nee, M. & Spooner, D.M. Solanaceae—a model for linking genomics with biodiversity. Comp. Funct. Genomics 5, 285–291 (2004).
Pnueli, L. et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989 (1998).
Lifschitz, E. & Eshed, Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57, 3405–3414 (2006).
Lippman, Z.B. et al. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 6, e288 (2008).
Child, A. A review of branching patterns in the Solanaceae. in The Biology and Taxonomy of the Solanaceae (eds. Hawkes, J.G., Lester, R.N. & Skelding, A.D.) 345–356 (Academic Press, London, 1979).
Danert, S. Die verzweigung der solanaceen in reproduktiven bereich. Abhandlungen der Deutschen Akademie der Wissenschaften zu Berlin 6, 1–183 (1958).
Rickett, H.W. The classification of inflorescences. Bot. Rev. 10, 187–231 (1944).
Park, S.J., Jiang, K., Schatz, M.C. & Lippman, Z.B. Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109, 639–644 (2012).
Lukyanenko, A.N., Ochova, E.P. & Egeyan, M. A mutant with a single flower terminating the main stem. TGC Report 23, 24 (1973).
Lifschitz, E. et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103, 6398–6403 (2006).
Poethig, R.S. Phase change and the regulation of developmental timing in plants. Science 301, 334–336 (2003).
Shalit, A. et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106, 8392–8397 (2009).
Cheng, X., Zhang, D., Cheng, Z., Keller, B. & Ling, H.-Q. Anew family of Ty1-copia-like retrotransposons originated in the tomato genome by a recent horizontal transfer event. Genetics 181, 1183–1193 (2009).
Xiao, H., Jiang, N., Schaffner, E., Stockinger, E.J. & van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530 (2008).
Menda, N., Semel, Y., Peled, D., Eshed, Y. & Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 38, 861–872 (2004).
Henikoff, S., Till, B.J. & Comai, L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636 (2004).
Zhao, L. et al. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. 37, 694–706 (2004).
Yoshida, A., Suzaki, T., Tanaka, W. & Hirano, H.Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 106, 20103–20108 (2009).
Takeda, S. et al. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 66, 1066–1077 (2011).
Cho, E. & Zambryski, P.C. ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc. Natl. Acad. Sci. USA 108, 2154–2159 (2011).
Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E. & Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000).
Uimari, A. et al. Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl. Acad. Sci. USA 101, 15817–15822 (2004).
Efroni, I., Blum, E., Goldshmidt, A. & Eshed, Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306 (2008).
Hempel, F.D. et al. Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124, 3845–3853 (1997).
Allen, K.D. & Sussex, I.M. Falsiflora and anantha control early stages of floral meristem development in tomato (Lycopersicon esculentum Mill.). Planta 200, 254–264 (1996).
William, D.A. et al. Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 101, 1775–1780 (2004).
Parcy, F., Nilsson, O., Busch, M.A., Lee, I. & Weigel, D. A genetic framework for floral patterning. Nature 395, 561–566 (1998).
Chae, E., Tan, Q.K.-G., Hill, T.A. & Irish, V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135, 1235–1245 (2008).
Souer, E. et al. Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20, 2033–2048 (2008).
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J.-I. & Nagato, Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180 (2012).
Weigel, D. & Nilsson, O. A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500 (1995).
Torti, S. et al. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24, 444–462 (2012).
Frijters, D. Principles of simulation of inflorescence development. Ann. Bot. 42, 549–560 (1978).
Prusinkiewicz, P., Erasmus, Y., Lane, B., Harder, L.D. & Coen, E. Evolution and development of inflorescence architectures. Science 316, 1452–1456 (2007).
Molinero-Rosales, N. et al. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 20, 685–693 (1999).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Liu, Y.-G. & Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–650 (2007).
McCallum, C.M., Comai, L., Greene, E.A. & Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 18, 455–457 (2000).
Eshed, Y., Baum, S.F. & Bowman, J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209 (1999).
Curtis, M.D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Van Eck, J., Kirk, D. & Walmsley, A. Tomato (Lycopersicum esculentum). Methods Mol. Biol. 343, 459–473 (2006).
Liang, D. et al. Establishment of a patterned GAL4-VP16 transactivation system for discovering gene function in rice. Plant J. 46, 1059–1072 (2006).
Ornitz, D.M., Moreadith, R.W. & Leder, P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc. Natl. Acad. Sci. USA 88, 698–702 (1991).
Fernandez, A.I. et al. Flexible tools for gene expression and silencing in tomato. Plant Physiol. 151, 1729–1740 (2009).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Néron, B. et al. Mobyle: a new full web bioinformatics framework. Bioinformatics 25, 3005–3011 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Blekhman, R., Marioni, J.C., Zumbo, P., Stephens, M. & Gilad, Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 20, 180–189 (2010).
R Development Core Team. A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2011).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57, 289–300 (1995).
Vojtek, A.B. & Hollenberg, S.M. RAS-RAF interaction: two-hybrid analysis. Methods Enzymol. 255, 331–342 (1995).
Bartel, P.L. et al. Using the two-hybrid system to detect protein-protein interactions. in Cellular Interactions in Development: A Practical Approach (ed. Hartley, D.A.) 153–179 (Oxford University Press, Oxford, 1993).
Fromont-Racine, M., Rain, J.-C. & Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277–282 (1997).
Formstecher, E. et al. Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 (2005).
Acknowledgements
We thank A. Aharoni for allowing screening of the population from which tmf2-D was derived. We also thank the Tomato Genetics Resource Center (TGRC) at the University of California, Davis, for germplasm, J. Van Eck for generating transgenic plants and DuPont Pioneer for funding. The work in the laboratory of A. Aharoni was supported by European Research Council (ERC) project SAMIT (Framework Programme 7). The tomato TILLING project is supported by funding from the European Union SOL Integrated Project FOOD-CT-2006-016214 to A.B. C.A.M. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation. This research was supported by research grant 1294-10 from the Israel Science Foundation and Binational Agricultural Research & Development Fund IS-4249-09 to Y.E. and a Career Development Award from The International Human Frontier Science Program Organization to Z.B.L.
Author information
Authors and Affiliations
Contributions
C.A.M., S.J.P. and K.J. designed and performed experiments. F.M., A.B. and Y.I. contributed reagents. C.A.M., Y.E. and Z.B.L. designed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–9 and Supplementary Table 4 (PDF 885 kb)
Supplementary Table 1
TMF interacting proteins (XLSX 12 kb)
Supplementary Table 2
Differentially expressed genes of tmf vs. WT (XLSX 86 kb)
Supplementary Table 3
Primer sequences (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
MacAlister, C., Park, S., Jiang, K. et al. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet 44, 1393–1398 (2012). https://doi.org/10.1038/ng.2465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2465
This article is cited by
-
Screening and identification of photoresponse factors in kiwifruit (Actinidia arguta) development
Molecular Biology Reports (2024)
-
The genome of Aechmea fasciata provides insights into the evolution of tank epiphytic habits and ethylene-induced flowering
Communications Biology (2022)
-
The auxin-responsive transcription factor SlDOF9 regulates inflorescence and flower development in tomato
Nature Plants (2022)
-
SISTER OF TM3 activates FRUITFULL1 to regulate inflorescence branching in tomato
Horticulture Research (2021)
-
The APETALA2 homolog CaFFN regulates flowering time in pepper
Horticulture Research (2021)