Abstract
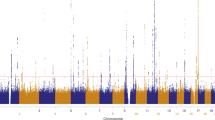
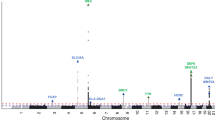
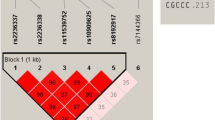
We previously reported a genome-wide association study (GWAS) identifying 14 susceptibility loci for generalized vitiligo. We report here a second GWAS (450 individuals with vitiligo (cases) and 3,182 controls), an independent replication study (1,440 cases and 1,316 controls) and a meta-analysis (3,187 cases and 6,723 controls) identifying 13 additional vitiligo-associated loci. These include OCA2-HERC2 (combined P = 3.80 × 10−8), MC1R (P = 1.82 × 10−13), a region near TYR (P = 1.57 × 10−13), IFIH1 (P = 4.91 × 10−15), CD80 (P = 3.78 × 10−10), CLNK (P = 1.56 × 10−8), BACH2 (P = 2.53 × 10−8), SLA (P = 1.58 × 10−8), CASP7 (P = 3.56 × 10−8), CD44 (P = 1.78 × 10−9), IKZF4 (P = 2.75 × 10−14), SH2B3 (P = 3.54 × 10−18) and TOB2 (P = 6.81 × 10−10). Most vitiligo susceptibility loci encode immunoregulatory proteins or melanocyte components that likely mediate immune targeting and the relationships among vitiligo, melanoma, and eye, skin and hair coloration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Picardo, M. & Taïeb, A. (eds.). Vitiligo (Springer, Heidelberg & New York, 2010).
Alkhateeb, A. et al. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 16, 208–214 (2003).
Jin, Y. et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 362, 1686–1697 (2010).
Jin, Y. et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat. Genet. 42, 576–578 (2010).
Birlea, S.A. et al. Comprehensive association analysis of candidate genes for generalized vitiligo supports XBP1, FOXP3, and TSLP. J. Invest. Dermatol. 131, 371–381 (2011).
Jin, Y. et al. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J. Investig. Dermatol. published online, doi:10.1038/jid.2012.37 (8 March 2012).
Spritz, R.A. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2, 78 (2010).
Rinchik, E.M. et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361, 72–76 (1993).
Kayser, M. et al. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am. J. Hum. Genet. 82, 411–423 (2008).
Sturm, R.A. et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 82, 424–431 (2008).
Eiberg, H. et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 123, 177–187 (2008).
Jannot, A.-S. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur. J. Hum. Genet. 13, 913–920 (2005).
Amos, C.I. et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 20, 5012–5023 (2011).
Cook, A.L. et al. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J. Invest. Dermatol. 129, 392–405 (2009).
Skipper, J.C. et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 183, 527–534 (1996).
Touloukian, C.E., Leitner, W.W., Robbins, P.F., Rosenberg, S. & Restifo, N.P. Mining the melanosome for tumor vaccine targets: P polypeptide is a novel tumor-associated antigen. Cancer Res. 61, 8100–8104 (2001).
Tomany, S.C., Klein, R. & Klein, B.E.K. The relationship between iris color, hair color, and skin sun sensitivity and the 10-year incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 110, 1526–1533 (2003).
Duffy, D.L. et al. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am. J. Hum. Genet. 80, 241–252 (2007).
Sulem, P. et al. Genetic determinants of hair, eye, and skin pigmentation in Europeans. Nat. Genet. 39, 1443–1452 (2007).
Dessinioti, C., Antoniou, C., Katsambas, A. & Stratigos, A.J. Melanocortin 1 receptor variants: functional role and pigmentary associations. Photochem. Photobiol. 87, 978–987 (2011).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006).
Smyth, D.J. et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 38, 617–619 (2006).
Sutherland, A. et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. J. Clin. Endocrinol. Metab. 92, 3338–3341 (2007).
Martínez, A. et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur. J. Hum. Genet. 16, 861–864 (2008).
Li, Y. et al. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J. Invest. Dermatol. 130, 2768–2772 (2010).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233 (2009).
Peach, R.J. et al. Both extracellular immunoglobin-like domains of CD80 contain residues critical for binding T cell surface receptors CTLA-4 and CD28. J. Biol. Chem. 270, 21181–21187 (1995).
Stamper, C.C. et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature 410, 608–611 (2001).
Wu, J.N. & Koretzky, G.A. The SLP-76 family of adapter proteins. Semin. Immunol. 16, 379–393 (2004).
Sasaki, S. et al. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene 19, 3739–3749 (2000).
Cooper, J.D. et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40, 1399–1401 (2008).
Grant, S.F. et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes 58, 290–295 (2009).
Dubois, P.C. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Dragone, L.L., Shaw, L.A., Myers, M.D. & Weiss, A. SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol. Rev. 232, 218–228 (2009).
Tomer, Y. & Greenberg, D. The thyroglobulin gene as the first thyroid-specific susceptibility gene for autoimmune thyroid disease. Trends Mol. Med. 10, 306–308 (2004).
Lamkanfi, M. & Kanneganti, T.D. Caspase-7: a protease involved in apoptosis and inflammation. Int. J. Biochem. Cell Biol. 42, 21–24 (2010).
García-Lozano, J.R. et al. Caspase 7 influences susceptibility to rheumatoid arthritis. Rheumatology (Oxford) 46, 1243–1247 (2007).
Babu, S.R. et al. Caspase 7 is a positional candidate gene for IDDM 17 in a Bedouin Arab family. Ann. NY Acad. Sci. 1005, 340–343 (2003).
Baaten, B.J., Li, C.R. & Bradley, L.M. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 3, 508–512 (2010).
Ramos, P.S. et al. Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 63, 2049–2057 (2011).
Pan, F. et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146 (2009).
Hakonarson, H. et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes 57, 1143–1146 (2008).
Petukhova, L. et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117 (2010).
Devallière, J. & Charreau, B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem. Pharmacol. 82, 1391–1402 (2011).
Smyth, D.J. et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 359, 2767–2777 (2008).
Hunt, K.A. et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 40, 395–402 (2008).
Coenen, M.J. et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum. Mol. Genet. 18, 4195–4203 (2009).
Alcina, A. et al. The autoimmune disease-associated KIF5A, CD226 and SH2B3 gene variants confer susceptibility for multiple sclerosis. Genes Immun. 11, 439–445 (2010).
Jia, S. & Meng, A. Tob genes in development and homeostasis. Dev. Dyn. 236, 913–921 (2007).
Seya, T., Matsumoto, M., Ebihara, T. & Oshiumi, H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol. Rev. 227, 44–53 (2009).
Taïeb, A. & Picardo, M. The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res. 20, 27–35 (2007).
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Li, Y., Willer, C.J., Ding, J., Scheet, P. & Abecasis, G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Yang, J. et al. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Falconer, D.S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 29, 51–76 (1965).
Risch, N. Assessing the role of HLA-linked and unlinked determinants of disease. Am. J. Hum. Genet. 40, 1–14 (1987).
Howitz, J., Brodthagen, H., Schwartz, M. & Thompsen, K. Prevalence of vitiligo. Epidemiological survey on the isle of Bornholm, Denmark. Arch. Dermatol. 113, 47–52 (1977).
Szklarczyk, D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39 Database Issue, D561–D568 (2011).
Acknowledgements
We thank the many patients with vitiligo and healthy control individuals around the world who participated in this study. We thank the University of Colorado Cancer Center Genomics and Microarray Core for genome-wide genotyping and BodySync (Aurora, Colorado, USA) for replication genotyping. This work was supported by grants R01AR045584, R01AR056292 and P30AR057212 from the U.S. National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Y.J. performed statistical analyses. K.G. managed computer databases and genotype data. T.M.F., S.B., S.A.B., S.L.R. and J.B.C. managed DNA samples and contributed to experimental procedures. P.J.H. managed subject coordination. S.A.B., D.C.B., R.M.L., A.W., J.P.W.v.d.V., M.R.W., W.T.M., E.H.K., D.J.G., A.P.W., M.P., G.L., A.T., T.J., K.E., N.v.G., J.L., A.O., A.H., S.E., G.S. and N.B.S. provided subject samples and phenotype information. P.R.F. and R.A.S. oversaw and managed all aspects of the study. R.A.S. wrote the first draft of the manuscript. All authors contributed to the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–4. (PDF 1379 kb)
Rights and permissions
About this article
Cite this article
Jin, Y., Birlea, S., Fain, P. et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet 44, 676–680 (2012). https://doi.org/10.1038/ng.2272
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2272
This article is cited by
-
Vitiligo Treatments: Review of Current Therapeutic Modalities and JAK Inhibitors
American Journal of Clinical Dermatology (2023)
-
A mouse model of vitiligo based on endogenous auto-reactive CD8 + T cell targeting skin melanocyte
Cell Regeneration (2022)
-
A comprehensive meta-analysis and prioritization study to identify vitiligo associated coding and non-coding SNV candidates using web-based bioinformatics tools
Scientific Reports (2022)
-
Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors
Cellular & Molecular Immunology (2021)
-
Association of GZMB polymorphisms and susceptibility to non-segmental vitiligo in a Korean population
Scientific Reports (2021)