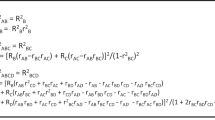Abstract
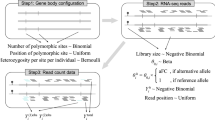
RNA profiling can be used to capture the expression patterns of many genes that are associated with expression quantitative trait loci (eQTLs). Employing published putative cis eQTLs, we developed a Bayesian approach to predict SNP genotypes that is based only on RNA expression data. We show that predicted genotypes can accurately and uniquely identify individuals in large populations. When inferring genotypes from an expression data set using eQTLs of the same tissue type (but from an independent cohort), we were able to resolve 99% of the identities of individuals in the cohort at Padjusted ≤ 1 × 10−5. When eQTLs derived from one tissue were used to predict genotypes using expression data from a different tissue, the identities of 90% of the study subjects could be resolved at Padjusted ≤ 1 × 10−5. We discuss the implications of deriving genotypic information from RNA data deposited in the public domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Beer, D.G. et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8, 816–824 (2002).
Hoshida, Y. et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 359, 1995–2004 (2008).
Barrett, T. et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37, D885–D890 (2009).
Parkinson, H. et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 35, D747–D750 (2007).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Dimas, A.S. et al. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science 325, 1246–1250 (2009).
Greenawalt, D.M. et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011).
Schadt, E.E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008).
Schadt, E.E. et al. Genetics of gene expression surveyed in maize, mouse and man. Nature 422, 297–302 (2003).
Smith, E.N. & Kruglyak, L. Gene-environment interaction in yeast gene expression. PLoS Biol. 6, e83 (2008).
Yang, X. et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 41, 415–423 (2009).
Hertzberg, L. et al. Prediction of chromosomal aneuploidy from gene expression data. Genes Chromosom. Cancer 46, 75–86 (2007).
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Hao, K., Chudin, E., Greenawalt, D. & Schadt, E.E. Magnitude of stratification in human populations and impacts on genome wide association studies. PLoS ONE 5, e8695 (2010).
Tan, P.K. et al. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 31, 5676–5684 (2003).
Baumbusch, L.O. et al. Comparison of the Agilent, ROMA/NimbleGen and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics 9, 379 (2008).
Barnes, M., Freudenberg, J., Thompson, S., Aronow, B. & Pavlidis, P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 33, 5914–5923 (2005).
Lamb, J.R. et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS ONE 6, e20090 (2011).
Wang, S.M., Ooi, L.L. & Hui, K.M. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin. Cancer Res. 13, 6275–6283 (2007).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Heid, I.M. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960 (2010).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Musunuru, K. et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 (2010).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Yang, X. et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 20, 1020–1036 (2010).
Zhong, H. et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 6, e1000932 (2010).
Zhong, H., Yang, X., Kaplan, L.M., Molony, C. & Schadt, E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 86, 581–591 (2010).
Lanktree, M.B. et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 88, 6–18 (2011).
Couzin-Frankel, J. Ethics. DNA returned to tribe, raising questions about consent. Science 328, 558 (2010).
Author information
Authors and Affiliations
Contributions
E.E.S., S.W. and K.H. designed the methods and experiments, and jointly analyzed the data sets. E.E.S. and K.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–3 and Supplementary Note (PDF 274 kb)
Rights and permissions
About this article
Cite this article
Schadt, E., Woo, S. & Hao, K. Bayesian method to predict individual SNP genotypes from gene expression data. Nat Genet 44, 603–608 (2012). https://doi.org/10.1038/ng.2248
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2248
This article is cited by
-
Sequre: a high-performance framework for secure multiparty computation enables biomedical data sharing
Genome Biology (2023)
-
Ethical implications of epigenetics in the era of personalized medicine
Clinical Epigenetics (2022)
-
Dysregulated expression levels of APH1B in peripheral blood are associated with brain atrophy and amyloid-β deposition in Alzheimer’s disease
Alzheimer's Research & Therapy (2021)
-
Flimma: a federated and privacy-aware tool for differential gene expression analysis
Genome Biology (2021)
-
Recovering genotypes and phenotypes using allele-specific genes
Genome Biology (2021)