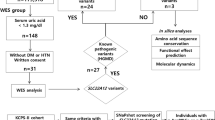
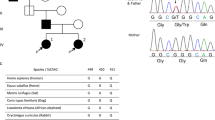
Abstract
Familial hyperkalemic hypertension (FHHt) is a Mendelian form of arterial hypertension that is partially explained by mutations in WNK1 and WNK4 that lead to increased activity of the Na+-Cl− cotransporter (NCC) in the distal nephron. Using combined linkage analysis and whole-exome sequencing in two families, we identified KLHL3 as a third gene responsible for FHHt. Direct sequencing of 43 other affected individuals revealed 11 additional missense mutations that were associated with heterogeneous phenotypes and diverse modes of inheritance. Polymorphisms at KLHL3 were not associated with blood pressure. The KLHL3 protein belongs to the BTB-BACK-kelch family of actin-binding proteins that recruit substrates for Cullin3-based ubiquitin ligase complexes. KLHL3 is coexpressed with NCC and downregulates NCC expression at the cell surface. Our study establishes a role for KLHL3 as a new member of the complex signaling pathway regulating ion homeostasis in the distal nephron and indirectly blood pressure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
22 March 2012
In the version of this article initially published, two references were omitted, resulting in several statements being incorrectly attributed in the Online Methods. In the 'Ped01' subsection of the 'Whole-exome sequencing' section, two statements were attributed to ref. 21. The correct reference for these statements has been added as ref. 27. In the 'Ped02' subsection of the 'Whole-exome sequencing' section, Annovar was incorrectly attributed to ref. 22. The correct reference has been added as ref. 28. As a result of the addition of these two references, former refs. 27-36 have been renumbered as refs. 29-38, respectively, in the text and reference list. In addition, author affiliation 7 was incorrectly given as Genes and Disease Program, Center for Genomic Regulation (CGR), Pompeu Fabra University, Barcelona, Spain. The correct affiliation is Genes and Disease Program, Center for Genomic Regulation (CGR) and Pompeu Fabra University, Barcelona, Spain. Similarly, affiliation 32 was incorrectly given as Genomic and Epigenetic Variation in Disease Group, Center for Genomic Regulation, Universitat Pompeu Fabra, Barcelona, Spain. The correct affiliation is Genomic and Epigenetic Variation in Disease Group, CGR and Pompeu Fabra University, Barcelona, Spain. These corrections have been made in the HTML and PDF versions of the article.
References
Lifton, R.P., Gharavi, A.G. & Geller, D.S. Molecular mechanisms of human hypertension. Cell 104, 545–556 (2001).
Meneton, P., Jeunemaitre, X., de Wardener, H.E. & MacGregor, G.A. Links between dietary salt intake, renal salt handling, blood pressure, and cardiovascular diseases. Physiol. Rev. 85, 679–715 (2005).
Gordon, R.D. et al. Gordon's syndrome: a sodium-volume-dependent form of hypertension with a genetic basis. in Hypertension: Pathophysiology, Diagnosis and Management (eds. Laragh, J.H. & Brenner, B.M.) 2111–2113 (Raven Press, 1995).
Wilson, F.H. et al. Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 (2001).
Lalioti, M.D. et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat. Genet. 38, 1124–1132 (2006).
Yang, S.S. et al. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4D561A/+ knockin mouse model. Cell Metab. 5, 331–344 (2007).
Hadchouel, J., Delaloy, C., Faure, S., Achard, J.M. & Jeunemaitre, X. Familial hyperkalemic hypertension. J. Am. Soc. Nephrol. 17, 208–217 (2006).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Charru, A., Jeunemaitre, X., Soubrier, F., Corvol, P. & Chatellier, G. HYPERGENE: a clinical and genetic database for genetic analysis of human hypertension. J. Hypertens. 12, 981–985 (1994).
Lai, F. et al. Molecular characterization of KLHL3, a human homologue of the Drosophila kelch gene. Genomics 66, 65–75 (2000).
Soltysik-Espanola, M. et al. Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol. Biol. Cell 10, 2361–2375 (1999).
Kigoshi, Y., Tsuruta, F. & Chiba, T. Ubiquitin ligase activity of Cul3-KLHL7 protein Is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J. Biol. Chem. 286, 33613–33621 (2011).
Adams, J., Kelso, R. & Cooley, L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10, 17–24 (2000).
Prag, S., De Arcangelis, A., Georges-Labouesse, E. & Adams, J.C. Regulation of post-translational modifications of muskelin by protein kinase C. Int. J. Biochem. Cell Biol. 39, 366–378 (2007).
Kelso, R.J., Hudson, A.M. & Cooley, L. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J. Cell Biol. 156, 703–713 (2002).
Friedman, P.A. & Gesek, F.A. Stimulation of calcium transport by amiloride in mouse distal convoluted tubule cells. Kidney Int. 48, 1427–1434 (1995).
Frindt, G. & Palmer, L.G. Surface expression of sodium channels and transporters in rat kidney: effects of dietary sodium. Am. J. Physiol. Renal Physiol. 297, F1249–F1255 (2009).
Hamm-Alvarez, S.F. & Sheetz, M.P. Microtubule-dependent vesicle transport: modulation of channel and transporter activity in liver and kidney. Physiol. Rev. 78, 1109–1129 (1998).
Pintard, L., Willems, A. & Peter, M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687 (2004).
Arroyo, J.P. et al. Nedd4–2 modulates renal Na+-Cl− cotransporter via the aldosterone-SGK1-Nedd4–2 pathway. J. Am. Soc. Nephrol. 22, 1707–1719 (2011).
Ko, B. et al. RasGRP1 stimulation enhances ubiquitination and endocytosis of the sodium-chloride cotransporter. Am. J. Physiol. Renal Physiol. 299, F300–F309 (2010).
Boyden, L.M. et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482, 98–102 (2012).
Ulgen, A. & Li, W. Comparing single-nucleotide polymorphism marker-based and microsatellite marker-based linkage analyses. BMC Genet. 6 ( suppl. 1, S13 (2005).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Sobel, E. & Lange, K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58, 1323–1337 (1996).
Lindner, T.H. & Hoffmann, K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 21, 405–407 (2005).
Bonnefond, A. et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome.. PLoS One 5, e13630 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data.. Nucleic Acids Res. 38, e164 (2010).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ng, P.C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Li, B. et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 (2009).
Ehret, G.B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Sali, A., Potterton, L., Yuan, F., van Vlijmen, H. & Karplus, M. Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 (1995).
Cheval, L. et al. Atlas of gene expression in the mouse kidney: new features of glomerular parietal cells. Physiol. Genomics 43, 161–173 (2011).
Richardson, C. et al. Activation of the thiazide-sensitive Na+-Cl− cotransporter by the WNK-regulated kinases SPAK and OSR1. J. Cell Sci. 121, 675–684 (2008).
Del Nery, E. et al. Rab6A and Rab6A' GTPases play non-overlapping roles in membrane trafficking. Traffic 7, 394–407 (2006).
Loïodice, I. et al. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 15, 3333–3344 (2004).
Acknowledgements
We thank C. Büsst for her critical reading of the manuscript, E. Clauser, F. Auradé and C. Auzan for helpful discussions, E. Durand, S. Lecointe and F. De Graeve for their contribution to whole-exome sequencing and M. Longépée-Dupas for assistance in family screening. This work was supported by INSERM, the Agence Nationale pour la Recherche (ANR; 05-MRAR-010-01), the European Union Framework Programme 7 through the HYPERGENE project (HEALTH-F4-2007-201550) and through the GEUVADIS project (HEALTH-261123), the Leducq Foundation (Transatlantic Network on Hypertension; 07 CVD 01), the Fondation Leducq Trans-Atlantic Network of Excellence (05 CVD 01; Preventing Sudden Death), the Groupe de Réflexion sur la Recherche Cardiovasculaire and Biotronik and the Ministry of Science and Innovation (MICINN; SAF2008-00357).
Author information
Authors and Affiliations
Consortia
Contributions
H.L.-D.-P. designed, performed and analyzed the data of the genetic analyses and immunoblotting experiments, and wrote the corresponding parts of the manuscript. J.B. designed, performed and analyzed the data of the genetic analyses (Ped02). D.T. together with S.O. and X.E. designed, performed and analyzed the WES of the Ped02 kindred. S.M.-L. designed and performed the in vitro and immunofluorescence experiments, and wrote the corresponding parts of the manuscript. N.B.-N. designed the WES for the Ped01 kindred and participated in the interpretation of the genetic analyses (exome sequencing of Ped01 and association studies) and in the writing of the manuscript. O.P. designed, performed and analyzed KLHL3 modeling and the prediction of the consequences of KLHL3 mutations. G.B. participated in the collection of the families with FHHt and to the analysis of previous candidate genes. V.E. collected and characterized the first members of the Nantes FHHt family. A.B. contributed to the WES of Ped01. O.S. performed the bioinformatics analyses of WES in Ped01. C. Simian performed the sequencing of hypertensive and normotensive individuals. E.V.-P. designed, performed and analyzed the qRT-PCR experiments. C. Soukaseum provided technical help to H.L.-D.-P. C. Mandet performed the immunohistochemistry. F.B., O.C., M.D., B.F., P.H., J.K., M.K., P.L., C. Mourani, P.N., V.P., C.T., R.J.U., S.D.S. and C.I.B. recruited families with FHHt. G.E. and M.C. designed, performed and analyzed the association study of KLHL3 and blood pressure within the ICBP. P.B. designed and analyzed the immunohistochemistry experiments. P.F. designed and analyzed WES. J.-J.S. designed and analyzed the genetic study and wrote the corresponding parts of the manuscript. X.J. and J.H. designed the study, analyzed data and co-wrote the manuscript. X.J. organized the entire study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–3, 5 and 6 and Supplementary Note (PDF 21357 kb)
Supplementary Table 4
Association of common variants at the KLHL3 locus with blood pressure in healthy individuals (XLS 255 kb)
Rights and permissions
About this article
Cite this article
Louis-Dit-Picard, H., Barc, J., Trujillano, D. et al. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44, 456–460 (2012). https://doi.org/10.1038/ng.2218
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2218
This article is cited by
-
Insights into the diverse mechanisms and effects of variant CUL3-induced familial hyperkalemic hypertension
Cell Communication and Signaling (2023)
-
Familial hyperkalemic hypertension: hyperkalemia not hypertension defines dominant KLHL3 disease and may permit earlier recognition and tailored therapy
Journal of Nephrology (2022)
-
Genotype–phenotype correlation in Gordon’s syndrome: report of two cases carrying novel heterozygous mutations
Journal of Nephrology (2022)
-
Kelch-like protein 3 in human disease and therapy
Molecular Biology Reports (2022)
-
Monogenic forms of low-renin hypertension: clinical and molecular insights
Pediatric Nephrology (2022)