Abstract
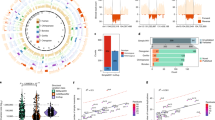
Using comparative sequencing approaches, we investigated the evolutionary history of the European-enriched 17q21.31 MAPT inversion polymorphism. We present a detailed, BAC-based sequence assembly of the inverted human H2 haplotype and compare it to the sequence structure and genetic variation of the corresponding 1.5-Mb region for the noninverted H1 human haplotype and that of chimpanzee and orangutan. We found that inversion of the MAPT region is similarly polymorphic in other great ape species, and we present evidence that the inversions occurred independently in chimpanzees and humans. In humans, the inversion breakpoints correspond to core duplications with the LRRC37 gene family. Our analysis favors the H2 configuration and sequence haplotype as the likely great ape and human ancestral state, with inversion recurrences during primate evolution. We show that the H2 architecture has evolved more extensive sequence homology, perhaps explaining its tendency to undergo microdeletion associated with mental retardation in European populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
NCBI Reference Sequence
Change history
17 August 2008
NOTE: In the version of this article initially published online, the left side of Figure 4c was mislabeled. The error has been corrected for the print, PDF and HTML versions of this article.
References
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004).
Tuzun, E., Bailey, J.A. & Eichler, E.E. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 14, 493–506 (2004).
Chimpanzee Sequence and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005).
Cheng, Z. et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93 (2005).
Stefansson, H. et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005).
Gijselinck, I. et al. Visualization of MAPT inversion on stretched chromosomes of tau-negative frontotemporal dementia patients. Hum. Mutat. 27, 1057–1059 (2006).
Cruts, M. et al. Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the tau region. Hum. Mol. Genet. 14, 1753–1762 (2005).
Baker, M. et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 8, 711–715 (1999).
Pittman, A.M. et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum. Mol. Genet. 13, 1267–1274 (2004).
Myers, A.J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–1499 (2007).
Conrad, C. et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann. Neurol. 41, 277–281 (1997).
Rademakers, R. et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum. Mol. Genet. 14, 3281–3292 (2005).
Sundar, P.D. et al. Two sites in the MAPT region confer genetic risk for Guam ALS/PDC and dementia. Hum. Mol. Genet. 16, 295–306 (2007).
Myers, A.J. et al. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum. Mol. Genet. 14, 2399–2404 (2005).
Sharp, A.J. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 38, 1038–1042 (2006).
Koolen, D.A. et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 38, 999–1001 (2006).
Shaw-Smith, C. et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 38, 1032–1037 (2006).
Evans, W. et al. The tau H2 haplotype is almost exclusively Caucasian in origin. Neurosci. Lett. 369, 183–185 (2004).
Hardy, J. et al. Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem. Soc. Trans. 33, 582–585 (2005).
Bailey, J.A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Johnson, M.E. et al. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc. Natl. Acad. Sci. USA 103, 17626–17631 (2006).
Caceres, M., Sullivan, R.T. & Thomas, J.W. A recurrent inversion on the eutherian X chromosome. Proc. Natl. Acad. Sci. USA 104, 18571–18576 (2007).
Ebersberger, I. et al. Mapping human genetic ancestry. Mol. Biol. Evol. 24, 2266–2276 (2007).
Gagneux, P. The genus Pan: population genetics of an endangered outgroup. Trends Genet. 18, 327–330 (2002).
Yu, N. et al. Low nucleotide diversity in chimpanzees and bonobos. Genetics 164, 1511–1518 (2003).
Kehrer-Sawatzki, H., Sandig, C.A., Goidts, V. & Hameister, H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet. Genome Res. 108, 91–97 (2005).
Rosenberg, N.A. et al. Genetic structure of human populations. Science 298, 2381–2385 (2002).
Jakobsson, M. et al. Genotype, haplotype, and copy number variation in worldwide human populations. Nature 451, 998–1003 (2008).
Zody, M.C. et al. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature 440, 671–675 (2006).
Jiang, Z. et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (2007).
Cardone, M.F. et al. Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 9, R28 (2008).
Casals, F. & Navarro, A. Chromosomal evolution: inversions: the chicken or the egg? Heredity 99, 479–480 (2007).
Lupski, J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998).
Osborne, L.R. et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 29, 321–325 (2001).
Gimelli, G. et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Mol. Genet. 12, 849–858 (2003).
Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J. & Eichler, E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017 (2001).
Parsons, J. Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci. 11, 615–619 (1995).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Higgins, D.G. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 25, 307–318 (1994).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Goodman, M. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64, 31–39 (1999).
Acknowledgements
We thank T. Marques, D. Reich, A. Navarro, N. Patterson, S. McCarroll, T. Brown and K. Augustyn for critical comments and valuable discussions in the preparation of this manuscript; C. Alkan for providing computational assistance; and members of the Broad Institute Sequence Platform (BISP) and Washington University Genome Sequencing Center (WUGSC) for generating clone-based sequencing data for this project. M.C.Z., A.A., W.C.W., L.W.H., T.A.G., R.K.W., A.D.R., J.W., J.G. and the BISP and WUGSC were supported by grants from the National Human Genome Research Institute. This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project number Z01 AG000957-05. This work was supported by NIH grants GM058815 and HG002385 to E.E.E. and a Rosetta Inpharmatics Fellowship (Merck Laboratories) to Z.J. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Z.J., M.C.Z. and E.E.E. analyzed and annotated sequence organization; J.M.K. did the haplotype analyses; F.A., M.F.C. and M.V. developed the FISH inversion assay and typed all primate metaphase chromosomes; L.C. and Z.C. did the segmental duplication analyses; M.C.Z., W.C.W., A.A., T.A.G., L.W.H., A.D.R., H.C.F., J.W., J.G. and J.D. generated, sequenced and analyzed the BAC clone assembly; R.K.W. oversaw sequence production; E.E.E., M.C.Z., Z.J., W.C.W., J.H. and J.M.K. drafted the manuscript; W.C.W., J.H. and E.E.E. designed the study; and E.E.E. finalized the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1 and 2, Supplementary Tables 1–5 (PDF 7487 kb)
Rights and permissions
About this article
Cite this article
Zody, M., Jiang, Z., Fung, HC. et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 40, 1076–1083 (2008). https://doi.org/10.1038/ng.193
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.193
This article is cited by
-
Discovering genetic mechanisms underlying the co-occurrence of Parkinson’s disease and non-motor traits
npj Parkinson's Disease (2024)
-
17q21.31 sub-haplotypes underlying H1-associated risk for Parkinson’s disease are associated with LRRC37A/2 expression in astrocytes
Molecular Neurodegeneration (2022)
-
Sex-specific recombination patterns predict parent of origin for recurrent genomic disorders
BMC Medical Genomics (2021)
-
Tau haplotypes support the Asian ancestry of the Roma population settled in the Basque Country
Heredity (2018)
-
Approaches and advances in the genetic causes of autoimmune disease and their implications
Nature Immunology (2018)