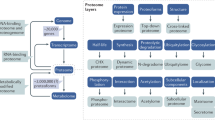
Abstract
Knowledge of the human genome has fertilized research in the embryonic field of proteomics. The aim of this Review is to examine the recent application of emerging proteomic technologies to diagnosis of renal disease. We discuss the roles, efficacy and diagnostic potential of different proteomic approaches, focusing on current difficulties and potential solutions. Our rudimentary knowledge of the healthy human urine proteome is described, as are studies that have sought to use the urinary proteome as a tool for diagnosis of renal disease. Vignettes of renal proteome are also presented. The integral role of bioinformatics, and the need for standardized sample preservation and reporting of results, are discussed.
Key Points
-
Urine can harbor proteins from all compartments of the kidney; the urinary proteome therefore has the potential to act as a diagnostic and prognostic indicator in patients with renal disease
-
Application of new techniques to mapping of the healthy urinary proteome continues to advance elucidation of this crucial 'baseline' information
-
The wide concentration range of proteins and peptides in urine is a barrier to effective proteomic analysis
-
Proteomic analysis of uremic toxins from plasma, and from renal cells and tissues, also holds promise
-
Despite recent advances in proteomic technologies, the full clinical utility of this type of analysis in nephrology is far from being realized
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Aebersold R and Mann M (2003) Mass spectrometry-based proteomics. Nature 422: 198–207
Hachey DL and Chaurand P (2004) Proteomics in reproductive medicine: the technology for separation and identification of proteins. J Reprod Immunol 63: 61–73
Hirsch J et al. (2004) Proteomics: current techniques and potential applications to lung disease. Am J Physiol Lung Cell Mol Physiol 287: L1–L23
Gygi SP and Aebersold R (2000) Mass spectrometry and proteomics. Curr Opin Chem Biol 4: 489–494
Issaq HJ (2001) The role of separation science in proteomics research. Electrophoresis 22: 3629–3638
Venter JC et al. (2001) The sequence of the human genome. Science 291: 1304–1351
Lander ES et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921
Anderson NL et al. (2004) The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics 3: 311–326
Marshall AG et al. (1998) Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev 17: 1–35
Ibarrola N et al. (2003) A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem 75: 6043–6049
Liu T et al. (2005) Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res 4: 2070–2080
Hao G et al. (2006) SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA 103: 1012–1017
Ong SE and Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1: 252–262
Hewitt SM et al. (2004) Discovery of protein biomarkers for renal diseases. J Am Soc Nephrol 15: 1677–1689
Tencer J et al. (1994) Stability of albumin, protein HC, immunoglobulin G, kappa- and lambda-chain immunoreactivity, orosomucoid and alpha 1-antitrypsin in urine stored at various conditions. Scand J Clin Lab Invest 54: 199–206
Serafini-Cessi F et al. (2003) Tamm-Horsfall glycoprotein: biology and clinical relevance. Am J Kidney Dis 42: 658–676
Zolotarjova N et al. (2005) Differences among techniques for high-abundant protein depletion. Proteomics 5: 3304–3313
Marshall T and Williams KM (1986) Electrophoresis indicates protein loss on centrifugation of urine. Clin Chem 32: 2105–2106
Schaub S et al. (2004) Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol 15: 219–227
Schaub S et al. (2004) Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int 65: 323–332
O'Riordan E et al. (2004) Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol 15: 3240–3248
Schaub S et al. (2005) Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am J Transplant 5: 729–738
Jackson DW et al. (1993) Altered urinary excretion of lysosomal hydrolases in pregnancy. Am J Kidney Dis 22: 649–655
Taracha E et al. (2004) Alanine aminopeptidase activity in urine: a new marker of chronic alcohol abuse? Alcohol Clin Exp Res 28: 729–735
Osicka TM et al. (2000) Albuminuria in patients with type 1 diabetes is directly linked to changes in the lysosome-mediated degradation of albumin during renal passage. Diabetes 49: 1579–1584
Issaq HJ et al. (2005) Multidimensional separation of peptides for effective proteomic analysis. J Chromatogr B Analyt Technol Biomed Life Sci 817: 35–47
Liu H et al. (2002) Multidimensional separations for protein/peptide analysis in the post-genomic era. Biotechniques 32: 898, 900, 902 passim
Cooper JW et al. (2004) Recent advances in capillary separations for proteomics. Electrophoresis 25: 3913–3926
Giddings J (1987) Concepts and comparisons in multidimensional separation. J High Resolut Chromatogr Commun 10: 319–323
O'Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250: 4007–4021
Edwards JJ et al. (1982) Proteins of human urine. III. Identification and two-dimensional electrophoretic map positions of some major urinary proteins. Clin Chem 28: 941–948
Griffith OW and Meister A (1979) Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J Biol Chem 254: 7558–7560
Brookes PS et al. (2002) High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics 2: 969–977
Mann M et al. (2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 20: 261–268
Weissinger EM et al. (2004) Proteomic patterns established with capillary electrophoresis and mass spectrometry for diagnostic purposes. Kidney Int 65: 2426–2434
Fung E et al. (2003) The use of SELDI protein chip array technology in renal disease research. In Renal Disease: Techniques and Protocols, 295–314 (Ed Goligorsky MS) Totowa: Humana Press
Wang Y (2004) Immunostaining with dissociable antibody microarrays. Proteomics 4: 20–26
Arthur JM (2003) Proteomics. Curr Opin Nephrol Hypertens 12: 423–430
Gygi SP et al. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999
DeSouza L et al. (2005) Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res 4: 377–386
Kiernan UA et al. (2003) Comparative urine protein phenotyping using mass spectrometric immunoassay. J Proteome Res 2: 191–197
Claverie J-M (2003) Bioinformatics for Dummies. New York: Wiley
Elias JE et al. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2: 667–675
Li J et al. (2002) Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 48: 1296–1304
Adam BL et al. (2002) Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res 62: 3609–3614
Granger CB et al. (2004) National Heart, Lung, And Blood Institute Clinical Proteomics Working Group report. Circulation 109: 1697–1703
Oh J et al. (2004) Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics 4: 3485–3497
Pieper R et al. (2004) Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics 4: 1159–1174
Smith G et al. (2005) Development of a high-throughput method for preparing human urine for two-dimensional electrophoresis. Proteomics 5: 2315–2318
Lafitte D et al. (2002) Optimized preparation of urine samples for two-dimensional electrophoresis and initial application to patient samples. Clin Biochem 35: 581–589
Thongboonkerd V et al. (2002) Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int 62: 1461–1469
Celis JE et al. (1999) A comprehensive protein resource for the study of bladder cancer: http://biobase.dk/cgi-bin/celis. Electrophoresis 20: 300–309
Spahr CS et al. (2001) Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 1: 93–107
Pisitkun T et al. (2004) Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101: 13368–13373
Norden AG et al. (2004) Quantitative amino acid and proteomic analysis: very low excretion of polypeptides >750 Da in normal urine. Kidney Int 66: 1994–2003
Cutillas PR et al. (2004) The urinary proteome in Fanconi syndrome implies specificity in the reabsorption of proteins by renal proximal tubule cells. Am J Physiol Renal Physiol 287: F353–F364
Clarke W et al. (2003) Characterization of renal allograft rejection by urinary proteomic analysis. Ann Surg 237: 660–664
Weissinger EM et al. (2004) Proteomics: a novel tool to unravel the patho-physiology of uraemia. Nephrol Dial Transplant 19: 3068–3077
Mischak H et al. (2004) Proteomic analysis for the assessment of diabetic renal damage in humans. Clin Sci (Lond) 107: 485–495
Haubitz M et al. (2005) Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int 67: 2313–2320
Hampel DJ et al. (2001) Toward proteomics in uroscopy: urinary protein profiles after radiocontrast medium administration. J Am Soc Nephrol 12: 1026–1035
Cadieux PA et al. (2004) Surface-enhanced laser desorption/ionization-time of flight-mass spectrometry (SELDI-TOF-MS): a new proteomic urinary test for patients with urolithiasis. J Clin Lab Anal 18: 170–175
Rogers MA et al. (2003) Proteomic profiling of urinary proteins in renal cancer by surface enhanced laser desorption ionization and neural-network analysis: identification of key issues affecting potential clinical utility. Cancer Res 63: 6971–6983
Thongboonkerd V et al. (2004) Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol 15: 650–662
Kwapiszewska G et al. (2004) Identification of proteins in laser-microdissected small cell numbers by SELDI-TOF and Tandem MS. BMC Biotechnol 4: 30
Chaurand P et al. (2004) Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am J Pathol 165: 1057–1068
Bagshaw RD et al. (2005) A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol Cell Proteomics 4: 133–143
Molina H et al. (2005) A proteomic analysis of human hemodialysis fluid. Mol Cell Proteomics 4: 637–650
Carr S et al. (2004) The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol Cell Proteomics 3: 531–533
Orchard S et al. (2005) Further steps towards data standardisation: the Proteomic Standards Initiative HUPO 3rd annual congress, Beijing 25-27th October, 2004. Proteomics 5: 337–339
Hanash S (2004) HUPO initiatives relevant to clinical proteomics. Mol Cell Proteomics 3: 298–301
Marouga R et al. (2005) The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem 382: 669–678
Campostrini N et al. (2005) Spot overlapping in two-dimensional maps: a serious problem ignored for much too long. Proteomics 5: 2385–2395
Cho A and Normile D (2002) Nobel Prize in Chemistry. Mastering macromolecules. Science 298: 527–528
Breiman L et al. (1984) Classification and Regression Trees. Belmont: Wadsworth International Group
Breiman L (2001) Random Forests. Machine Learning 45: 5–32
Qu Y et al. (2002) Boosted decision tree analysis of surface-enhanced laser desorption/ionization mass spectral serum profiles discriminates prostate cancer from noncancer patients. Clin Chem 48: 1835–1843
Woroniecki R et al. (2006) Urinary proteome of steroid-sensitive and steroid-resistant idiopathic nephrotic syndrome of childhood. Am J Nephrol 26: 258–267
Wagner M et al. (2004) Computational protein biomarker prediction: a case study for prostate cancer. BMC Bioinformatics 5: 26
Yu JK et al. (2004) An integrated approach to the detection of colorectal cancer utilizing proteomics and bioinformatics. World J Gastroenterol 10: 3127–3131
Vapnik V (1995) The Nature of Statistical Learning Theory. New York: Springer
Ball G et al. (2002) An integrated approach utilizing artificial neural networks and SELDI mass spectrometry for the classification of human tumours and rapid identification of potential biomarkers. Bioinformatics 18: 395–404
Acknowledgements
The authors' studies were supported in part by NIH grants DK45462, DK54602, DK52783 (MSG) and Westchester Artificial Kidney Foundation. E O'Riordan is a recipient of the Kevin J and Gloria B Kiely National Kidney Foundation Fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
O'Riordan, E., Gross, S. & Goligorsky, M. Technology Insight: renal proteomics—at the crossroads between promise and problems. Nat Rev Nephrol 2, 445–458 (2006). https://doi.org/10.1038/ncpneph0241
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncpneph0241
This article is cited by
-
Application of proteomic analysis to the study of renal diseases
Nature Reviews Nephrology (2009)