Abstract
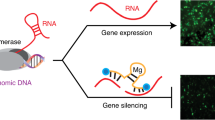
Synthetic molecules that recognize specific sequences within cellular DNA are potentially powerful tools for investigating chromosome structure and function. Here, we designed antigene peptide nucleic acids (agPNAs) to target the transcriptional start sites for the human progesterone receptor B (hPR-B) and A (hPR-A) isoforms at sequences predicted to be single-stranded within the open complex of chromosomal DNA. We found that the agPNAs were potent inhibitors of transcription, showing for the first time that synthetic molecules can recognize transcription start sites inside cells. Breast cancer cells treated with agPNAs showed marked changes in morphology and an unexpected relationship between the strictly regulated levels of hPR-B and hPR-A. We confirmed these phenotypes using siRNAs and antisense PNAs, demonstrating the power of combining antigene and antisense strategies for gene silencing. agPNAs provide a general approach for controlling transcription initiation and a distinct option for target validation and therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kaihatsu, K., Janowski, B.A. & Corey, D.R. Recognition of chromosomal DNA by PNAs. Chem. Biol. 11, 749–758 (2004).
Nielsen, P.E., Egholm, M., Berg, R.H. & Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine substituted polyamide. Science 254, 1497–1500 (1991).
Egholm, M. et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365, 566–568 (1993).
Demidov, V.V., Broude, N.E., Lavrentieva-Smolina, I.V., Kuhn, H. & Frank-Kamenetskii, M.D. An artificial primosome: design, function, and applications. ChemBioChem 2, 133–139 (2001).
Larsen, H.J. & Nielsen, P.E. Transcription induced binding of peptide nucleic acid (PNA) to double-stranded DNA: sequence-specific suicide transcription. Nucleic Acids Res. 24, 458–463 (1996).
Mollegaard, N.E., Buchardt, O., Egholm, M. & Nielsen, P.E. Peptide nucleic acid-DNA strand displacement loops as artificial gene promoters. Proc. Natl. Acad. Sci. USA 91, 3892–3895 (1994).
Faruqi, A.F., Egholm, M. & Glazer, P.M. Peptide nucleic acid-targeted mutagenesis of chromosomal DNA. Proc. Natl. Acad. Sci. USA 95, 1398–1403 (1998).
Bentin, T. & Nielsen, P.E. Enhanced peptide nucleic acid binding to supercoiled DNA: possible implications for DNA breathing dynamics. Biochemistry 35, 8863–8869 (1996).
Zhang, X., Ishihara, T. & Corey, D.R. Strand invasion by PNAs and PNA-peptide chimera. Nucleic Acids Res. 28, 3332–3338 (2000).
Holstege, F.C., Fiedler, U. & Timmers, H.T. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16, 7468–7480 (1997).
Kahl, B.F., Li, H. & Paule, M.R. DNA melting and promoter clearance by eukaryotic RNA polymerase I. J. Mol. Biol. 299, 75–89 (2000).
Kastner, P. et al. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor isoforms A and B. EMBO J. 9, 1603–1614 (1990).
Misrahi, M. et al. Structure of the human progesterone receptor gene. Biochim. Biophys. Acta 1216, 289–292 (1993).
Conneely, O.M., Jericevic, B.M. & Lydon, J.P. Progesterone receptors in mammary gland development and tumorigenesis. J. Mammary Gland Biol. Neoplasia 8, 205–214 (2003).
Kaihatsu, K., Huffman, K.E. & Corey, D.R. Intracellular uptake and inhibition of gene expression by PNAs and PNA-peptide conjugates. Biochemistry 43, 14340–14347 (2004).
Herbert, B. et al. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. USA 96, 14726–14781 (1999).
Doyle, D.F., Braasch, D.A., Simmons, C.G., Janowski, B.A. & Corey, D.R. Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry 40, 53–64 (2001).
Liu, Y., Braasch, D.A., Nulf, C.J. & Corey, D.R. Isoform-specific inhibition of cellular gene expression by peptide nucleic acid. Biochemistry 43, 1921–1927 (2004).
Nulf, C.J. & Corey, D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs). Nucleic Acids Res. 32, 3792–3798 (2004).
Nardulli, A.M., Greene, G.L., O'Malley, B.W. & Katzenellenbogen, B.S. Regulation of progesterone receptor messenger ribonucleic acid and protein levels in MCF-7 cells by estradiol: analysis of estrogen's effect on progesterone receptor synthesis and degradation. Endocrinology 122, 935–944 (1988).
Vienonen, A., Syvala, H., Miettinen, S., Tuohimaa, P. & Ylikomi, T. Expression of progesterone receptor isoforms A and B is differentially regulated by estrogen in different breast cancer cell lines. J. Steroid Biochem. Mol. Biol. 80, 307–313 (2002).
Stein, C. Keeping the biotechnology of antisense in context. Nat. Biotechnol. 17, 209 (1999).
Jackson, A.L. & Linsley, P.S. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 20, 521–524 (2004).
Karmakar, S. & Das, C. Modulation of ezrin and E-cadherin expression by IL-1β and TGF-β1 in human trophoblasts. J. Reprod. Immunol. 64, 9–29 (2004).
Hunter, K.W. Ezrin, a key component in tumor metastasis. Trends Mol. Med. 10, 201–204 (2004).
Song, J. et al. Estradiol-induced ezrin overexpression in ovarian cancer: a new signaling domain for estrogen. Cancer Lett. 220, 57–65 (2005).
Condon, J.C. et al. A decline in the levels of progesterone receptor coactivators in the pregnant uterus at term may antagonize progesterone receptor function and contribute to the initiation of parturition. Proc. Natl. Acad. Sci. USA 100, 9518–9523 (2003).
McGowan, E.M. et al. Cytoskeletal responsiveness to progestins is dependent on progesterone receptor A levels. J. Mol. Endocrinol. 31, 241–253 (2003).
Milne, L., Xu, Y., Perrin, D.M. & Sigman, D.S. An approach to gene-specific transcription inhibition using oligonucleotides complementary to the template strand of the open complex. Proc. Natl. Acad. Sci. USA 97, 3136–3141 (2000).
Knauert, M.P. & Glazer, P.M. Triplex forming oligonucleotides:sequence-specific tools for gene targeting. Hum. Mol. Genet. 10, 2243–2251 (2001).
Besch, R., Giovannangeli, C., Schuh, T., Kammerbauer, C. & Degitz, K. Characterization and quantification of triple helix formation in chromosomal DNA. J. Mol. Biol. 341, 979–989 (2004).
Dervan, P.B. & Edelson, B.S. Recognition of the DNA minor groove by pyrrole-imidizole polyamides. Curr. Opin. Struct. Biol. 13, 284–299 (2003).
Dudouet, B. et al. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 10, 859–867 (2003).
Janowski, B.A. et al. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. (in the press) (2005).
Mattick, J.S. & Makunin, I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 14, R121–R132 (2005).
Acknowledgements
This work was supported by the US National Institutes of Health (NIGMS 60642 and 73042 to D.R.C. and P01 HD011149 to C.R.M.) and the Robert A. Welch Foundation (I-1244).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Effect of estradiol treatment on the ability of agPNAs to inhibit hPR gene expression. (PDF 282 kb)
Rights and permissions
About this article
Cite this article
Janowski, B., Kaihatsu, K., Huffman, K. et al. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat Chem Biol 1, 210–215 (2005). https://doi.org/10.1038/nchembio724
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio724
This article is cited by
-
Peptide nucleic acid-zirconium coordination nanoparticles
Scientific Reports (2023)
-
Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions
Nature Communications (2016)
-
Delivery of cell-penetrating peptide-peptide nucleic acid conjugates by assembly on an oligonucleotide scaffold
Scientific Reports (2015)
-
Antisense Oligonucleotides: Treating Neurodegeneration at the Level of RNA
Neurotherapeutics (2013)
-
Chemically Modified Oligonucleotides Modulate an Epigenetically Varied and Transient Form of Transcription Silencing of HIV-1 in Human Cells
Molecular Therapy - Nucleic Acids (2012)