Abstract
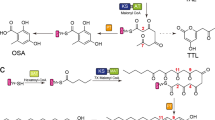
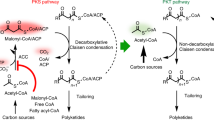
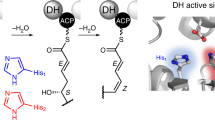
Iterative highly reducing polyketide synthases from filamentous fungi are the most complex and enigmatic type of polyketide synthase discovered to date. Here we uncover an unusual degree of programming by the hypothemycin highly reducing polyketide synthase, in which a single ketoreductase domain shows stereospecificity that is controlled by substrate length. Mapping of the structural domains responsible for this feature allowed for the biosynthesis of an unnatural diastereomer of the natural product dehydrozearalenol.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Weissman, K.J. & Leadlay, P.F. Nat. Rev. Microbiol. 3, 925–936 (2005).
Smith, S. & Tsai, S.C. Nat. Prod. Rep. 24, 1041–1072 (2007).
Cox, R.J. Org. Biomol. Chem. 5, 2010–2026 (2007).
Ma, S.M. et al. Science 326, 589–592 (2009).
Meier, J.L. & Burkart, M.D. Chem. Soc. Rev. 38, 2012–2045 (2009).
Oppermann, U. et al. Chem. Biol. Interact. 143–144, 247–253 (2003).
Siskos, A.P. et al. Chem. Biol. 12, 1145–1153 (2005).
Baerga-Ortiz, A. et al. Chem. Biol. 13, 277–285 (2006).
O'Hare, H.M., Baerga-Ortiz, A., Popovic, B., Spencer, J.B. & Leadlay, P.F. Chem. Biol. 13, 287–296 (2006).
Castonguay, R., He, W., Chen, A.Y., Khosla, C. & Cane, D.E. J. Am. Chem. Soc. 129, 13758–13769 (2007).
Castonguay, R. et al. J. Am. Chem. Soc. 130, 11598–11599 (2008).
Valenzano, C.R., Lawson, R.J., Chen, A.Y., Khosla, C. & Cane, D.E. J. Am. Chem. Soc. 131, 18501–18511 (2009).
Caffrey, P. ChemBioChem 4, 654–657 (2003).
Reid, R. et al. Biochemistry 42, 72–79 (2003).
Keatinge-Clay, A.T. & Stroud, R.M. Structure 14, 737–748 (2006).
Keatinge-Clay, A.T. Chem. Biol. 14, 898–908 (2007).
Zheng, J., Taylor, C.A., Piasecki, S.K. & Keatinge-Clay, A.T. Structure 18, 913–922 (2010).
Zhou, H. et al. J. Am. Chem. Soc. 132, 4530–4531 (2010).
Reeves, C.D., Hu, Z., Reid, R. & Kealey, J.T. Appl. Environ. Microbiol. 74, 5121–5129 (2008).
Celmer, W.D. J. Am. Chem. Soc. 87, 1801–1802 (1965).
Rossmann, M.G. & Argos, P. Annu. Rev. Biochem. 50, 497–532 (1981).
Filling, C. et al. J. Biol. Chem. 277, 25677–25684 (2002).
Hagler, W.M. et al. Appl. Environ. Microbiol. 37, 849–853 (1979).
Fisch, K.M. et al. J. Am. Chem. Soc. 133, 16635–16641 (2011).
Maier, T., Leibundgut, M. & Ban, N. Science 321, 1315–1322 (2008).
Acknowledgements
This work was supported by US National Institutes of Health grant 1R01GM085128 to Y.T., the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chair in Bioorganic and Medicinal Chemistry to J.C.V. We thank C. Reeves (Amyris Inc.) for the hpm genes.
Author information
Authors and Affiliations
Contributions
H.Z., Y.T. and J.C.V. conceived of the idea and designed the study. Z.G. and H.Z. performed the syntheses of all the thioester substrates in this study. H.Z. designed and performed molecular cloning. H.Z., K.Q. and J.W. performed the heterologous protein expression and purification as well as in vitro and in vivo characterization of the megasynthases. All authors analyzed and discussed the results. H.Z., Y.T. and J.C.V. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results and Supplementary Methods (PDF 2065 kb)
Rights and permissions
About this article
Cite this article
Zhou, H., Gao, Z., Qiao, K. et al. A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat Chem Biol 8, 331–333 (2012). https://doi.org/10.1038/nchembio.912
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.912
This article is cited by
-
Substrate control in stereoselective lanthionine biosynthesis
Nature Chemistry (2015)
-
Recent progress regarding the bioactivities, biosynthesis and synthesis of naturally occurring resorcinolic macrolides
Acta Pharmacologica Sinica (2014)
-
Strategies for mining fungal natural products
Journal of Industrial Microbiology and Biotechnology (2014)
-
Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides
Journal of Industrial Microbiology and Biotechnology (2014)
-
Human amniotic epithelial cells can differentiate into granulosa cells and restore folliculogenesis in a mouse model of chemotherapy-induced premature ovarian failure
Stem Cell Research & Therapy (2013)