Abstract
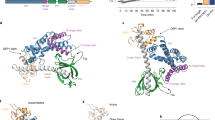
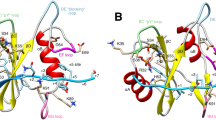
MAP kinases regulate essential cellular events, including cell growth, differentiation and inflammation. The solution structure of a complete MAPK–MAPK-regulatory protein complex, p38α–HePTP, was determined, enabling a comprehensive investigation of the molecular basis of specificity and fidelity in MAPK regulation. Structure determination was achieved by combining NMR spectroscopy and small-angle X-ray scattering data with a new ensemble calculation–refinement procedure. We identified 25 residues outside of the HePTP kinase interaction motif necessary for p38α recognition. The complex adopts an extended conformation in solution and rarely samples the conformation necessary for kinase deactivation. Complex formation also does not affect the N-terminal lobe, the activation loop of p38α or the catalytic domain of HePTP. Together, these results show how the downstream tyrosine phosphatase HePTP regulates p38α and provide for fundamentally new insights into MAPK regulation and specificity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Raman, M., Chen, W. & Cobb, M.H. Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 (2007).
Kim, E.K. & Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 1802, 396–405 (2010).
Cobb, M.H. et al. Regulation of the MAP kinase cascade. Cell. Mol. Biol. Res. 40, 253–256 (1994).
Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004).
Wilson, K.P. et al. Crystal structure of p38 mitogen-activated protein kinase. J. Biol. Chem. 271, 27696–27700 (1996).
Kallunki, T. et al. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8, 2996–3007 (1994).
Akella, R., Min, X., Wu, Q., Gardner, K.H. & Goldsmith, E.J. The third conformation of p38α MAP kinase observed in phosphorylated p38α and in solution. Structure 18, 1571–1578 (2010).
Chang, C.I., Xu, B.E., Akella, R., Cobb, M.H. & Goldsmith, E.J. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9, 1241–1249 (2002).
Heo, Y.S. et al. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 23, 2185–2195 (2004).
Liu, S., Sun, J.P., Zhou, B. & Zhang, Z.Y. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc. Natl. Acad. Sci. USA 103, 5326–5331 (2006).
Ma, W. et al. Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase. Structure 18, 1502–1511 (2010).
Reményi, A., Good, M.C., Bhattacharyya, R.P. & Lim, W.A. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell 20, 951–962 (2005).
Zhou, T., Sun, L., Humphreys, J. & Goldsmith, E.J. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure 14, 1011–1019 (2006).
Critton, D.A., Tortajada, A., Stetson, G., Peti, W. & Page, R. Structural basis of substrate recognition by hematopoietic tyrosine phosphatase. Biochemistry 47, 13336–13345 (2008).
Saxena, M., Williams, S., Brockdorff, J., Gilman, J. & Mustelin, T. Inhibition of T cell signaling by mitogen-activated protein kinase-targeted hematopoietic tyrosine phosphatase (HePTP). J. Biol. Chem. 274, 11693–11700 (1999).
Muñoz, J.J., Tarrega, C., Blanco-Aparicio, C. & Pulido, R. Differential interaction of the tyrosine phosphatases PTP-SL, STEP and HePTP with the mitogen-activated protein kinases ERK1/2 and p38α is determined by a kinase specificity sequence and influenced by reducing agents. Biochem. J. 372, 193–201 (2003).
Tanoue, T., Adachi, M., Moriguchi, T. & Nishida, E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2, 110–116 (2000).
Zhang, J., Zhou, B., Zheng, C.F. & Zhang, Z.Y. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 278, 29901–29912 (2003).
Zanke, B. et al. Cloning and expression of an inducible lymphoid-specific, protein tyrosine phosphatase (HePTPase). Eur. J. Immunol. 22, 235–239 (1992).
Lombroso, P.J., Murdoch, G. & Lerner, M. Molecular characterization of a protein-tyrosine-phosphatase enriched in striatum. Proc. Natl. Acad. Sci. USA 88, 7242–7246 (1991).
Hendriks, W., Schepens, J., Brugman, C., Zeeuwen, P. & Wieringa, B. A novel receptor-type protein tyrosine phosphatase with a single catalytic domain is specifically expressed in mouse brain. Biochem. J. 305, 499–504 (1995).
Saxena, M., Williams, S., Tasken, K. & Mustelin, T. Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nat. Cell Biol. 1, 305–311 (1999).
Bellon, S., Fitzgibbon, M.J., Fox, T., Hsiao, H.M. & Wilson, K.P. The structure of phosphorylated p38γ is monomeric and reveals a conserved activation-loop conformation. Structure 7, 1057–1065 (1999).
Diskin, R., Engelberg, D. & Livnah, O. A novel lipid binding site formed by the MAP kinase insert in p38α. J. Mol. Biol. 375, 70–79 (2008).
Masterson, L.R. et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat. Chem. Biol. 6, 821–828 (2010).
Masterson, L.R., Mascioni, A., Traaseth, N.J., Taylor, S.S. & Veglia, G. Allosteric cooperativity in protein kinase A. Proc. Natl. Acad. Sci. USA 105, 506–511 (2008).
Zhang, J. et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463, 501–506 (2010).
Piserchio, A. et al. Solution NMR insights into docking interactions involving inactive ERK2. Biochemistry 50, 3660–3672 (2011).
Vogtherr, M. et al. NMR backbone assignment of the mitogen-activated protein (MAP) kinase p38. J. Biomol. NMR 32, 175 (2005).
Esposito, V. et al. NMR assignment of the cAMP-binding domain A of the PKA regulatory subunit. J. Biomol. NMR 36 (suppl. 1), 64 (2006).
Vajpai, N. et al. Backbone NMR resonance assignment of the Abelson kinase domain in complex with imatinib. Biomol. NMR Assign. 2, 41–42 (2008).
Perry, J.J., Harris, R.M., Moiani, D., Olson, A.J. & Tainer, J.A. p38α MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses. J. Mol. Biol. 391, 1–11 (2009).
Hoffman, A.D., Urbatsch, I.L. & Vogel, P.D. Nucleotide binding to the human multidrug resistance protein 3, MRP3. Protein J. 29, 373–379 (2010).
Delannoy, S., Urbatsch, I.L., Tombline, G., Senior, A.E. & Vogel, P.D. Nucleotide binding to the multidrug resistance P-glycoprotein as studied by ESR spectroscopy. Biochemistry 44, 14010–14019 (2005).
Fitzgerald, C.E. et al. Structural basis for p38α MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity. Nat. Struct. Biol. 10, 764–769 (2003).
Vogtherr, M. et al. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew. Chem. Int. Edn Engl. 45, 993–997 (2006).
Kornev, A.P., Haste, N.M., Taylor, S.S. & Eyck, L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 103, 17783–17788 (2006).
Mustelin, T., Tautz, L. & Page, R. Structure of the hematopoietic tyrosine phosphatase (HePTP) catalytic domain: structure of a KIM phosphatase with phosphate bound at the active site. J. Mol. Biol. 354, 150–163 (2005).
Jeeves, M., McClelland, D.M., Barr, A.J. & Overduin, M. Sequence-specific 1H, 13C and 15N backbone resonance assignments of the 34 kDa catalytic domain of human PTPN7. Biomol. NMR Assign. 2, 101–103 (2008).
Gong, H. & Freed, K.F. Electrostatic solvation energy for two oppositely charged ions in a solvated protein system: salt bridges can stabilize proteins. Biophys. J. 98, 470–477 (2010).
Różycki, B., Kim, Y.C. & Hummer, G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure 19, 109–116 (2011).
Balasu, M.C. et al. Interface analysis of the complex between ERK2 and PTP-SL. PLoS ONE 4, e5432 (2009).
Peti, W. & Page, R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 51, 1–10 (2007).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Leonard, T.A., Rozycki, B., Saidi, L.F., Hummer, G. & Hurley, J.H. Crystal structure and allosteric activation of protein kinase C βII. Cell 144, 55–66 (2011).
Yang, S., Blachowicz, L., Makowski, L. & Roux, B. Multidomain assembled states of Hck tyrosine kinase in solution. Proc. Natl. Acad. Sci. USA 107, 15757–15762 (2010).
Franke, D. & Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Acknowledgements
The authors thank L. Yang and M. Allaire (National Synchrotron Light Source, NSLS) for their support at NSLS beamline X9. The authors thank P. Vogel (Southern Methodist University) for providing SL-ATP for the titration studies. This research was supported by grant RSG-08-067-01-LIB from the American Cancer Society to R.P. B.R. was supported by a Marie Curie International Outgoing Fellowship within the Seventh European Community Framework Program. B.R. and G.H. were supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health (NIH). Use of the NSLS at Brookhaven National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-98CH10886. 800 MHz NMR data were recorded at Brandeis University and were purchased with support from NIH S10-RR017269. Molecular simulations were performed on the Biowulf computing cluster at NIH.
Author information
Authors and Affiliations
Contributions
D.M.F. and D.K. cloned and purified all proteins. D.M.F. and W.P. performed and analyzed all NMR measurements. D.M.F., W.P. and R.P. performed and analyzed the SAXS measurements. D.K. performed all ITC measurements. R.P. and W.P. helped design the biochemical and NMR experiments. B.R. and G.H. performed all EROS and EROS-NMR calculations. R.P. and W.P. wrote the paper and all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 845 kb)
Rights and permissions
About this article
Cite this article
Francis, D., Różycki, B., Koveal, D. et al. Structural basis of p38α regulation by hematopoietic tyrosine phosphatase. Nat Chem Biol 7, 916–924 (2011). https://doi.org/10.1038/nchembio.707
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.707
This article is cited by
-
Mapping the deformability of natural and designed cellulosomes in solution
Biotechnology for Biofuels and Bioproducts (2022)
-
The expression patterns and the diagnostic/prognostic roles of PTPN family members in digestive tract cancers
Cancer Cell International (2020)
-
The methyl 13C-edited/13C-filtered transferred NOE for studying protein interactions with short linear motifs
Journal of Biomolecular NMR (2020)
-
1H, 15N and 13C sequence specific backbone assignment of the vanadate inhibited hematopoietic tyrosine phosphatase
Biomolecular NMR Assignments (2018)
-
Low potency toxins reveal dense interaction networks in metabolism
BMC Systems Biology (2016)