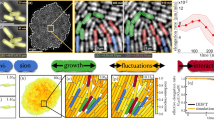
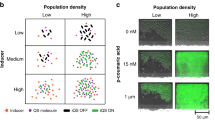
Abstract
It is postulated that in addition to cell density, other factors such as the dimensions and diffusional characteristics of the environment could influence quorum sensing (QS) and induction of genetic reprogramming. Modeling studies predict that QS may operate at the level of a single cell, but, owing to experimental challenges, the potential benefits of QS by individual cells remain virtually unexplored. Here we report a physical system that mimics isolation of a bacterium, such as within an endosome or phagosome during infection, and maintains cell viability under conditions of complete chemical and physical isolation. For Staphylococcus aureus, we show that quorum sensing and genetic reprogramming can occur in a single isolated organism. Quorum sensing allows S. aureus to sense confinement and to activate virulence and metabolic pathways needed for survival. To demonstrate the benefit of confinement-induced quorum sensing to individuals, we showed that quorum-sensing bacteria have significantly greater viability over non-QS bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
04 December 2009
In the version of this article initially published online, the funding support for C.J.B. was cited incorrectly. The error has been corrected for all versions of this article.
References
Fuqua, W., Winans, S. & Greenberg, E. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994).
Waters, C.M. & Bassler, B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005).
Diggle, S., Griffin, A., Campbell, G. & West, S. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414 (2007).
Hense, B. et al. Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239 (2007).
Fuqua, C., Parsek, M.R. & Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468 (2001).
Horswill, A.R., Stoodley, P., Stewart, P.S. & Parsek, M.R. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 387, 371–380 (2007).
Redfield, R. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10, 365–370 (2002).
Sandoz, K., Mitzimberg, S. & Schuster, M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. USA 104, 15876–15881 (2007).
Qazi, S.N.A. et al. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69, 7074–7082 (2001).
Alberghini, S. et al. Consequences of relative cellular positioning on quorum sensing and bacterial cell-to-cell communication. FEMS Microbiol. Lett. 292, 149–161 (2009).
James, S., Nilsson, P., James, G., Kjelleberg, S. & Fagerstrom, T. Luminescence control in the marine bacterium Vibrio fischeri: an analysis of the dynamics of lux regulation. J. Mol. Biol. 296, 1127–1137 (2000).
Dunman, P.M. et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353 (2001).
Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 (2003).
Shompole, S. et al. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Mol. Microbiol. 49, 919–927 (2003).
Baca, H.K. et al. Cell-directed assembly of lipid-silica nanostructures providing extended cell viability. Science 313, 337–341 (2006).
Lu, Y. et al. Aerosol-assisted self-assembly of mesostructured spherical nanoparticles. Nature 398, 223–226 (1999).
Murphy, R.F., Powers, S. & Cantor, C.R. Endosome pH measured in single cells by dual fluorescence flow cytometry: rapid acidification of insulin to pH 6. J. Cell Biol. 98, 1757–1762 (1984).
Baca, H.K. et al. Cell-directed assembly of bio/nano interfaces - a new scheme for cell immobilization. Acc. Chem. Res. 40, 836–845 (2007).
Jarry, T., Memmi, G. & Cheung, A. The expression of alpha-haemolysin is required for Staphylococcus aureus phagosomal escape after internalization in CFT-1 cells. Cell. Microbiol. 10, 1801–1814 (2008).
Novick, R.P. & Geisinger, E. Quorum sensing in Staphylococci. Annu. Rev. Genet. 42, 541–564 (2008).
Rothfork, J.M., Dessus-Babus, S., Van Wamel, W.J.B., Cheung, A.L. & Gresham, H.D. Fibrinogen depletion attenuates Staphyloccocus aureus infection by preventing density-dependent virulence gene UP-regulation. J. Immunol. 171, 5389–5395 (2003).
Peterson, M.M. et al. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4, 555–566 (2008).
Mathesius, U. et al. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 100, 1444–1449 (2003).
Shiner, E.K., Rumbaugh, K.P. & Williams, S.C. Interkingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol. Rev. 29, 935–947 (2005).
Balaban, N. et al. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187, 625–630 (2003).
Wang, R. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 (2007).
Rothfork, J.M. et al. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc. Natl. Acad. Sci. USA 101, 13867–13872 (2004).
Kupferwasser, L.I. et al. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Invest. 112, 222–233 (2003).
Doshi, D.A. et al. Optically, defined multifunctional patterning of photosensitive thin-film silica mesophases. Science 290, 107–111 (2000).
Acknowledgements
E.C.C. was supported by US Air Force Office of Scientific Research (AFOSR) grant FA 9550-07-1-0054 and US National Science Foundation Integrative Graduate Education and Research Traineeship Fellowship grant DGE-0504276. D.M.L. was supported by AFOSR grant FA 9550-07-1-0054 and Defense Threat Reduction Agency grant B0844671. N.P.D., A.C. and G.S.T. were supported by US National Institutes of Health (NIH) grants AI-037142 and AI-047441. H.G. was supported by NIH grant R01 AI-064926. C. J. B. was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering and Sandia National Laboratories’ Laboratory Directed Research and Development program. Further support was provided by the NIH/Roadmap for Medical Research grant PHS 2 PN2 EY016570B. The authors also acknowledge L. Kenney and M. Federle for useful comments. Some images in this paper were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility, supported as detailed at http://hsc.unm.edu/crtc/microscopy/Facility.html. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the US Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000.
Author information
Authors and Affiliations
Contributions
E.C.C. and D.M.L. conducted all of the experimental work on quorum sensing of isolated individual S. aureus. N.P.D. and A.C. constructed the GFP reporter strains. G.S.T. and H.G. conceived of testing quorum sensing of individual bacteria. C.J.B. conceived of the cell-directed assembly and aerosol-assisted self-assembly processes used for nanofabrication.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 530 kb)
Rights and permissions
About this article
Cite this article
Carnes, E., Lopez, D., Donegan, N. et al. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol 6, 41–45 (2010). https://doi.org/10.1038/nchembio.264
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.264
This article is cited by
-
Quorum sensing as a mechanism to harness the wisdom of the crowds
Nature Communications (2023)
-
High density lipoproteins mediate in vivo protection against staphylococcal phenol-soluble modulins
Scientific Reports (2021)
-
Bacteria-host transcriptional response during endothelial invasion by Staphylococcus aureus
Scientific Reports (2021)
-
Picking the right metaphors for addressing microbial systems: economic theory helps understanding biological complexity
International Microbiology (2021)
-
Single cyanobacteria@silica porous microcapsules via a sol–gel layer by layer for heavy-metal remediation
Journal of Sol-Gel Science and Technology (2019)