Abstract
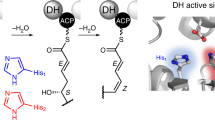
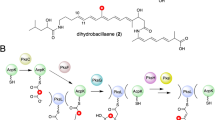
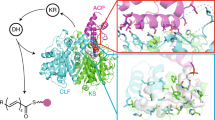
Modular polyketide synthases (PKSs) produce numerous structurally complex natural products that have diverse applications in medicine and agriculture. PKSs typically consist of several multienzyme subunits that utilize structurally defined docking domains (DDs) at their N and C termini to ensure correct assembly into functional multiprotein complexes. Here we report a fundamentally different mechanism for subunit assembly in trans-acyltransferase (trans-AT) modular PKSs at the junction between ketosynthase (KS) and dehydratase (DH) domains. This mechanism involves direct interaction of a largely unstructured docking domain (DD) at the C terminus of the KS with the surface of the downstream DH. Acyl transfer assays and mechanism-based crosslinking established that the DD is required for the KS to communicate with the acyl carrier protein appended to the DH. Two distinct regions for binding of the DD to the DH were identified using NMR spectroscopy, carbene footprinting, and mutagenesis, providing a foundation for future elucidation of the molecular basis for interaction specificity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009).
Keatinge-Clay, A.T. The structures of type I polyketide synthases. Nat. Prod. Rep. 29, 1050–1073 (2012).
Richter, C.D., Nietlispach, D., Broadhurst, R.W. & Weissman, K.J. Multienzyme docking in hybrid megasynthetases. Nat. Chem. Biol. 4, 75–81 (2008).
Weissman, K.J. & Müller, R. Protein-protein interactions in multienzyme megasynthetases. ChemBioChem 9, 826–848 (2008).
Fischbach, M.A. & Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Keatinge-Clay, A.T. Stereocontrol within polyketide assembly lines. Nat. Prod. Rep. 33, 141–149 (2016).
Wagner, D.T. et al. α-Methylation follows condensation in the gephyronic acid modular polyketide synthase. Chem. Commun. (Camb.) 52, 8822–8825 (2016).
Helfrich, E.J.N. & Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 33, 231–316 (2016).
Katz, L. The DEBS paradigm for type I modular polyketide synthases and beyond. Methods Enzymol. 459, 113–142 (2009).
Broadhurst, R.W., Nietlispach, D., Wheatcroft, M.P., Leadlay, P.F. & Weissman, K.J. The structure of docking domains in modular polyketide synthases. Chem. Biol. 10, 723–731 (2003).
Gokhale, R.S. & Khosla, C. Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol. 4, 22–27 (2000).
Buchholz, T.J. et al. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem. Biol. 4, 41–52 (2009).
Kuo, J., Lynch, S.R., Liu, C.W., Xiao, X. & Khosla, C. Partial in vitro reconstitution of an orphan polyketide synthase associated with clinical cases of nocardiosis. ACS Chem. Biol. 11, 2636–2641 (2016).
Gulder, T., Freeman, M. & Piel, J. The catalytic diversity of multimodular polyketide synthases: natural product biosynthesis beyond textbook assembly rules. Top. Curr. Chem. https://doi.org/10.1007/128_2010_113 (2011).
Dorival, J. et al. Characterization of intersubunit communication in the virginiamycin trans-acyl transferase polyketide synthase. J. Am. Chem. Soc. 138, 4155–4167 (2016).
Zeng, J. et al. Portability and structure of the four-helix bundle docking domains of trans-acyltransferase modular polyketide synthases. ACS Chem. Biol. 11, 2466–2474 (2016).
Song, L. et al. Discovery and biosynthesis of gladiolin: a Burkholderia gladioli antibiotic with promising activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 139, 7974–7981 (2017).
Eddy, S.R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
McGuffin, L.J., Bryson, K. & Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000).
Dosztányi, Z., Csizmok, V., Tompa, P. & Simon, I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 (2005).
Dosztányi, Z., Mészáros, B. & Simon, I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25, 2745–2746 (2009).
Worthington, A.S., Rivera, H., Torpey, J.W., Alexander, M.D. & Burkart, M.D. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem. Biol. 1, 687–691 (2006).
Kapur, S. et al. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg. Med. Chem. Lett. 18, 3034–3038 (2008).
Kosol, S., Contreras-Martos, S., Cedeño, C. & Tompa, P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules 18, 10802–10828 (2013).
Jensen, M.R., Ruigrok, R.W. & Blackledge, M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr. Opin. Struct. Biol. 23, 426–435 (2013).
Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 282, 1182–1189 (2015).
Manzi, L. et al. Carbene footprinting accurately maps binding sites in protein-ligand and protein-protein interactions. Nat. Commun. 7, 13288 (2016).
Pavlović-Lažetić, G.M. et al. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics 12, 66 (2011).
Vucetic, S. et al. DisProt: a database of protein disorder. Bioinformatics 21, 137–140 (2005).
Wright, P.E. & Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015).
Lin, Y.S., Hsu, W.L., Hwang, J.K. & Li, W.H. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol. Biol. Evol. 24, 1005–1011 (2007).
Nilsson, J., Grahn, M. & Wright, A.P.H. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 12, R65 (2011).
Bellay, J. et al. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 12, R14 (2011).
Neduva, V. & Russell, R.B. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 17, 465–471 (2006).
Gitlin, L., Hagai, T., LaBarbera, A., Solovey, M. & Andino, R. Rapid evolution of virus sequences in intrinsically disordered protein regions. PLoS Pathog. 10, e1004529 (2014).
Keatinge-Clay, A. Crystal structure of the erythromycin polyketide synthase dehydratase. J. Mol. Biol. 384, 941–953 (2008).
Wagner, D.T. et al. Structural and functional trends in dehydrating bimodules from trans-acyltransferase polyketide synthases. Structure 25, 1045–1055 e2 (2017).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Worthington, A.S. & Burkart, M.D. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org. Biomol. Chem. 4, 44–46 (2006).
Meier, J.L., Mercer, A.C., Rivera, H. Jr. & Burkart, M.D. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J. Am. Chem. Soc. 128, 12174–12184 (2006).
Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Lescop, E., Schanda, P. & Brutscher, B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 187, 163–169 (2007).
Palmer, A.G., III. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 104, 3623–3640 (2004).
Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013).
Schumann, F.H. et al. Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289 (2007).
Whitmore, L. & Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004).
Lees, J.G., Miles, A.J., Wien, F. & Wallace, B.A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962 (2006).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pierce, B.G. et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 (2014).
Acknowledgements
This research was supported by grants from the BBSRC (BB/L021692/1 to G.L.C. and BB/L022761/1 to J.R.L.). The Bruker MaXis II instrument used in this study was funded by the BBSRC (BB/M017982/1). N.J.O., J.E.M., L.M. and A.S.B. thank the University of Nottingham for funding. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement 639907 (to J.R.L.). G.L.C. is the recipient of a Wolfson Research Merit Award from the Royal Society (WM130033). We thank L. Song for assistance with LC-MS analyses.
Author information
Authors and Affiliations
Contributions
M.J. and G.L.C. designed the study, and all authors contributed to performing the research as follows: M.J. and D.G. carried out bioinformatics analyses; M.J. overproduced and purified all recombinant proteins, and conducted acyl transfer assays and LC–MS analyses of intact proteins; P.P. synthesized the trans-β-chloroacrylamide-containing pantetheine analog, and P.P. and M.J. carried out the crosslinking experiments; S.K. and J.R.L. conducted the NMR and CD experiments, and analyzed the data; A.S.B. and J.E.M. synthesized the diazirine reagent used in the footprinting experiments; L.M. and N.J.O. carried out the footprinting experiments and analyzed the data; M.J., S.K., J.R.L. and G.L.C. wrote the paper, and all authors contributed to revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figures 1–24 (PDF 4891 kb)
Rights and permissions
About this article
Cite this article
Jenner, M., Kosol, S., Griffiths, D. et al. Mechanism of intersubunit ketosynthase–dehydratase interaction in polyketide synthases. Nat Chem Biol 14, 270–275 (2018). https://doi.org/10.1038/nchembio.2549
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2549
This article is cited by
-
Diene incorporation by a dehydratase domain variant in modular polyketide synthases
Nature Chemical Biology (2022)
-
Aminoacyl chain translocation catalysed by a type II thioesterase domain in an unusual non-ribosomal peptide synthetase
Nature Communications (2022)
-
Structural basis for chain release from the enacyloxin polyketide synthase
Nature Chemistry (2019)