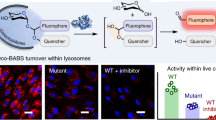
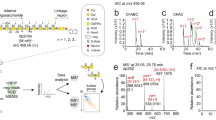
Abstract
Humans express at least two distinct β-glucuronidase enzymes that are involved in disease: exo-acting β-glucuronidase (GUSB), whose deficiency gives rise to mucopolysaccharidosis type VII, and endo-acting heparanase (HPSE), whose overexpression is implicated in inflammation and cancers. The medical importance of these enzymes necessitates reliable methods to assay their activities in tissues. Herein, we present a set of β-glucuronidase-specific activity-based probes (ABPs) that allow rapid and quantitative visualization of GUSB and HPSE in biological samples, providing a powerful tool for dissecting their activities in normal and disease states. Unexpectedly, we find that the supposedly inactive HPSE proenzyme proHPSE is also labeled by our ABPs, leading to surprising insights regarding structural relationships between proHPSE, mature HPSE, and their bacterial homologs. Our results demonstrate the application of β-glucuronidase ABPs in tracking pathologically relevant enzymes and provide a case study of how ABP-driven approaches can lead to discovery of unanticipated structural and biochemical functionality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Khan, F.I. et al. Large scale analysis of the mutational landscape in β-glucuronidase: a major player of mucopolysaccharidosis type VII. Gene 576, 36–44 (2016).
Sly, W.S., Quinton, B.A., McAlister, W.H. & Rimoin, D.L. Beta glucuronidase deficiency: report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J. Pediatr. 82, 249–257 (1973).
Naz, H. et al. Human β-glucuronidase: structure, function, and application in enzyme replacement therapy. Rejuvenation Res. 16, 352–363 (2013).
Rivara, S., Milazzo, F.M. & Giannini, G. Heparanase: a rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med. Chem. 8, 647–680 (2016).
Vlodavsky, I. et al. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 5, 115–132 (2012).
Cantarel, B.L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Jain, S. et al. Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 3, 375–381 (1996).
Wu, L., Viola, C.M., Brzozowski, A.M. & Davies, G.J. Structural characterization of human heparanase reveals insights into substrate recognition. Nat. Struct. Mol. Biol. 22, 1016–1022 (2015).
Jiang, J. et al. Detection of active mammalian GH31 α-glucosidases in health and disease using in-class, broad-spectrum activity-based probes. ACS Cent. Sci. 2, 351–358 (2016).
Kallemeijn, W.W. et al. Novel activity-based probes for broad-spectrum profiling of retaining β-exoglucosidases in situ and in vivo. Angew. Chem. Int. Ed. Engl. 51, 12529–12533 (2012).
Willems, L.I. et al. Potent and selective activity-based probes for GH27 human retaining α-galactosidases. J. Am. Chem. Soc. 136, 11622–11625 (2014).
Witte, M.D. et al. Ultrasensitive in situ visualization of active glucocerebrosidase molecules. Nat. Chem. Biol. 6, 907–913 (2010).
Jiang, J. et al. In vitro and in vivo comparative and competitive activity-based protein profiling of GH29 α-L-fucosidases. Chem. Sci. 6, 2782–2789 (2015).
Kwan, D.H. et al. Chemoenzymatic synthesis of 6-phospho-cyclophellitol as a novel probe of 6-phospho-β-glucosidases. FEBS Lett. 590, 461–468 (2016).
Barglow, K.T. & Cravatt, B.F. Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods 4, 822–827 (2007).
Li, K.Y. et al. Exploring functional cyclophellitol analogues as human retaining beta-glucosidase inhibitors. Org. Biomol. Chem. 12, 7786–7791 (2014).
Koshland, D.E. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. Camb. Philos. Soc. 28, 416–436 (1953).
Willems, L.I. et al. From covalent glycosidase inhibitors to activity-based glycosidase probes. Chemistry 20, 10864–10872 (2014).
Michikawa, M. et al. Structural and biochemical characterization of glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum. J. Biol. Chem. 287, 14069–14077 (2012).
Baici, A., Schenker, P., Wächter, M. & Rüedi, P. 3-Fluoro-2,4-dioxa-3-phosphadecalins as inhibitors of acetylcholinesterase. A reappraisal of kinetic mechanisms and diagnostic methods. Chem. Biodivers. 6, 261–282 (2009).
Speciale, G., Thompson, A.J., Davies, G.J. & Williams, S.J. Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr. Opin. Struct. Biol. 28, 1–13 (2014).
Gloster, T.M. & Davies, G.J. Glycosidase inhibition: assessing mimicry of the transition state. Org. Biomol. Chem. 8, 305–320 (2010).
Islam, M.R., Grubb, J.H. & Sly, W.S. C-terminal processing of human beta-glucuronidase. The propeptide is required for full expression of catalytic activity, intracellular retention, and proper phosphorylation. J. Biol. Chem. 268, 22627–22633 (1993).
Ono, M. et al. Phosphorylation of beta-glucuronidases from human normal liver and hepatoma by cAMP-dependent protein kinase. J. Biol. Chem. 263, 5884–5889 (1988).
Forrest, A.R. et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Weiss, D.J., Liggitt, D. & Clark, J.G. Histochemical discrimination of endogenous mammalian beta-galactosidase activity from that resulting from lac-Z gene expression. Histochem. J. 31, 231–236 (1999).
Peterson, S.B. & Liu, J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 32, 223–227 (2013).
Fairbanks, M.B. et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J. Biol. Chem. 274, 29587–29590 (1999).
Abboud-Jarrous, G. et al. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J. Biol. Chem. 283, 18167–18176 (2008).
Takahashi, K. et al. Characterization of CAA0225, a novel inhibitor specific for cathepsin L, as a probe for autophagic proteolysis. Biol. Pharm. Bull. 32, 475–479 (2009).
Shaw, E., Mohanty, S., Colic, A., Stoka, V. & Turk, V. The affinity-labelling of cathepsin S with peptidyl diazomethyl ketones. Comparison with the inhibition of cathepsin L and calpain. FEBS Lett. 334, 340–342 (1993).
Nadav, L. et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J. Cell Sci. 115, 2179–2187 (2002).
Vlodavsky, I. et al. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis 12, 112–127 (1992).
McKenzie, E. et al. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem. J. 373, 423–435 (2003).
Islam, M.R. et al. Active site residues of human beta-glucuronidase. Evidence for Glu(540) as the nucleophile and Glu(451) as the acid-base residue. J. Biol. Chem. 274, 23451–23455 (1999).
Kallemeijn, W.W. et al. A sensitive gel-based method combining distinct cyclophellitol-based probes for the identification of acid/base residues in human retaining β-glucosidases. J. Biol. Chem. 289, 35351–35362 (2014).
Niphakis, M.J. & Cravatt, B.F. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 83, 341–377 (2014).
Kawase, Y. et al. A-72363 A-1, A-2, and C, novel heparanase inhibitors from Streptomyces nobilis SANK 60192, II. Biological activities. J. Antibiot. (Tokyo) 49, 61–64 (1996).
Rabenstein, D.L. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 19, 312–331 (2002).
Davies, G.J., Wilson, K.S. & Henrissat, B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 (1997).
Bohlmann, L. et al. Functional and structural characterization of a heparanase. Nat. Chem. Biol. 11, 955–957 (2015).
Abboud-Jarrous, G. et al. Site-directed mutagenesis, proteolytic cleavage, and activation of human proheparanase. J. Biol. Chem. 280, 13568–13575 (2005).
Gingis-Velitski, S. et al. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J. Biol. Chem. 279, 44084–44092 (2004).
Moreland, R.J. et al. Lysosomal acid alpha-glucosidase consists of four different peptides processed from a single chain precursor. J. Biol. Chem. 280, 6780–6791 (2005).
Tollersrud, O.K. et al. Purification of bovine lysosomal alpha-mannosidase, characterization of its gene and determination of two mutations that cause alpha-mannosidosis. Eur. J. Biochem. 246, 410–419 (1997).
Li, K.-Y. Synthesis of cyclophellitol, dyclophellitol aziridine, and their tagged derivatives. European J. Org. Chem. 6030–6043 (2014).
Zheng, L., Baumann, U. & Reymond, J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115 (2004).
Li, M.Z. & Elledge, S.J. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852, 51–59 (2012).
Berger, I., Fitzgerald, D.J. & Richmond, T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 (2004).
Seiler, C.Y. et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 (2014).
Li, N. et al. Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat. Protoc. 8, 1155–1168 (2013).
D'Arcy, A., Bergfors, T., Cowan-Jacob, S.W. & Marsh, M. Microseed matrix screening for optimization in protein crystallization: what have we learned? Acta Crystallogr. F Struct. Biol. Commun. 70, 1117–1126 (2014).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Lebedev, A.A. et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 68, 431–440 (2012).
McNicholas, S., Potterton, E., Wilson, K.S. & Noble, M.E.M. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 (2011).
Acknowledgements
We thank the Diamond Light Source for access to beamlines i02, i03 and i04 (proposals mx-9948 and mx-13587), which contributed to the results presented here. We acknowledge the Netherlands Organization for Scientific Research (NWO, ChemThem Grant to J.M.F.G.A. and H.S.O.), the China Scholarship Council (CSC, PhD Grant to J.J.), the European Research Council (ErC-2011-AdG-290836 to H.S.O.; ErC-2012-AdG-322942 to G.J.D.), and the Royal Society (Ken Murray Research Professorship to G.J.D.) for financial support.
Author information
Authors and Affiliations
Contributions
L.W., J.M.F.G.A., H.S.O. and G.J.D. conceived and designed the experiments. J.J., M.A., W.D. and C.v.E. carried out synthesis of probes, with guidance from G.A.v.d.M. and J.D.C.C. L.W. and Y.J. carried out protein expression and structural studies on enzyme-probe complexes. J.J., L.W., W.W.K. and C.-L.K. carried out gel labeling experiments. J.J. and B.I.F. carried out proteomics experiments. C.-L.K. and W.W.K. determined IC50 and kinetic parameters for ABP inhibition. M.v.E. obtained tissue samples. L.W., J.J., H.S.O., and G.J.D. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–2, Supplementary Figures 1–12 and Supplementary Note 1 (PDF 4578 kb)
Supplementary Note 2
Synthesis and compound characterization details (PDF 6444 kb)
Supplementary Dataset 1
Full mass spectrometry datasets for proteomics experiments. (XLSX 2614 kb)
Rights and permissions
About this article
Cite this article
Wu, L., Jiang, J., Jin, Y. et al. Activity-based probes for functional interrogation of retaining β-glucuronidases. Nat Chem Biol 13, 867–873 (2017). https://doi.org/10.1038/nchembio.2395
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2395
This article is cited by
-
Chemoproteomic identification of a DPP4 homolog in Bacteroides thetaiotaomicron
Nature Chemical Biology (2023)
-
Microbial enzymes induce colitis by reactivating triclosan in the mouse gastrointestinal tract
Nature Communications (2022)
-
Indole bearing thiadiazole analogs: synthesis, β-glucuronidase inhibition and molecular docking study
BMC Chemistry (2019)