Abstract
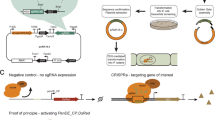
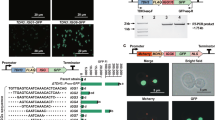
Here we report an efficient CRISPR–Cas9 knock-in strategy to activate silent biosynthetic gene clusters (BGCs) in streptomycetes. We applied this one-step strategy to activate multiple BGCs of different classes in five Streptomyces species and triggered the production of unique metabolites, including a novel pentangular type II polyketide in Streptomyces viridochromogenes. This potentially scalable strategy complements existing activation approaches and facilitates discovery efforts to uncover new compounds with interesting bioactivities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rutledge, P.J. & Challis, G.L. Nat. Rev. Microbiol. 13, 509–523 (2015).
Luo, Y., Cobb, R.E. & Zhao, H. Curr. Opin. Biotechnol. 30, 230–237 (2014).
Kang, H.S., Charlop-Powers, Z. & Brady, S.F. ACS Synth. Biol. 5, 1002–1010 (2016).
Cobb, R.E. & Zhao, H. Nat. Biotechnol. 30, 405–406 (2012).
Zhang, M.M., Wang, Y., Ang, E.L. & Zhao, H. Nat. Prod. Rep. 33, 963–987 (2016).
Doudna, J.A. & Charpentier, E. Science 346, 1258096 (2014).
Sander, J.D. & Joung, J.K. Nat. Biotechnol. 32, 347–355 (2014).
Cobb, R.E., Wang, Y. & Zhao, H. ACS Synth. Biol. 4, 723–728 (2015).
Huang, H., Zheng, G., Jiang, W., Hu, H. & Lu, Y. Acta Biochim. Biophys. Sin. (Shanghai) 47, 231–243 (2015).
Tong, Y., Charusanti, P., Zhang, L., Weber, T. & Lee, S.Y. ACS Synth. Biol. 4, 1020–1029 (2015).
Zeng, H. et al. Appl. Microbiol. Biotechnol. 99, 10575–10585 (2015).
Olano, C. et al. Microb. Biotechnol. 7, 242–256 (2014).
Bruheim, P., Sletta, H., Bibb, M.J., White, J. & Levine, D.W. J. Ind. Microbiol. Biotechnol. 28, 103–111 (2002).
Malpartida, F., Niemi, J., Navarrete, R. & Hopwood, D.A. Gene 93, 91–99 (1990).
Wang, W. et al. Appl. Environ. Microbiol. 79, 4484–4492 (2013).
Luo, Y., Zhang, L., Barton, K.W. & Zhao, H. ACS Synth. Biol. 4, 1001–1010 (2015).
Weber, T. et al. Nucleic Acids Res. 43, W237–W243 (2015).
Liu, W.T. et al. J. Antibiot. (Tokyo) 67, 99–104 (2014).
Luo, Y. et al. Nat. Commun. 4, 2894 (2013).
Moree, W.J. et al. ACS Chem. Biol. 9, 2300–2308 (2014).
Okuhara, M. et al. J. Antibiot. (Tokyo) 33, 13–17 (1980).
Eliot, A.C. et al. Chem. Biol. 15, 765–770 (2008).
Jomaa, H. et al. Science 285, 1573–1576 (1999).
Lackner, G. et al. J. Am. Chem. Soc. 129, 9306–9312 (2007).
Hillenmeyer, M.E., Vandova, G.A., Berlew, E.E. & Charkoudian, L.K. Proc. Natl. Acad. Sci. USA 112, 13952–13957 (2015).
Kieser, T., Bibb, M.J., Buttner, M.J., Chater, K.F. & Hopwood, D.A. Practical Streptomyces Genetics (John Innes Foundation, Norwich, UK, 2000).
Rhee, K.-H. & Davies, J. J. Microbiol. Biotechnol. 16, 1841–1848 (2006).
Acknowledgements
We gratefully acknowledge financial support from the National Institutes of Health (GM077596) (H.Z.), the A*STAR Visiting Investigator Program (H.Z.), and the National Research Foundation, Singapore (NRF2013-THE001-094) (M.M.Z., F.T.W., Y.H.L., E.L.A. and H.Z.). NMR data collection at the UIUC IGB Core was funded by NIH (S10-RR028833). Conjugative donor strains of E. coli were gifted by Prof. William Metcalf at UIUC. We thank members of the Zhao laboratory in UIUC, co-workers in MEL and MERL in A*STAR for constructive comments, Dr. Xudong Guan and Dr. Lingyang Zhu from UIUC for assisting with NMR data acquisition, and Dr. Ying Swan Ho from BTI, A*STAR for HRMS data acquisition.
Author information
Authors and Affiliations
Contributions
M.M.Z., F.T.W., and H.Z. conceived and designed the research. M.M.Z., F.T.W., Y.W., E.H., W.L.Y., R.E.C., and B.E. performed the molecular biology, conjugation and fermentation experiments. S.L. and Y.H.L. performed structure elucidation of compounds. M.M.Z., F.T.W., E.L.A., and H.Z. analyzed the data. M.M.Z., F.T.W., and H.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–8, Supplementary Figure 1–17 and Supplementary Note (PDF 3726 kb)
Rights and permissions
About this article
Cite this article
Zhang, M., Wong, F., Wang, Y. et al. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol 13, 607–609 (2017). https://doi.org/10.1038/nchembio.2341
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2341
This article is cited by
-
Bacillus and Streptomyces spp. as hosts for production of industrially relevant enzymes
Applied Microbiology and Biotechnology (2024)
-
Manipulation and epigenetic control of silent biosynthetic pathways in actinobacteria
World Journal of Microbiology and Biotechnology (2024)
-
Exploring a general multi-pronged activation strategy for natural product discovery in Actinomycetes
Communications Biology (2024)
-
Enhancing armeniaspirols production through multi-level engineering of a native Streptomyces producer
Microbial Cell Factories (2023)
-
Establishment of a visual gene knockout system based on CRISPR/Cas9 for the rare actinomycete Nonomuraea gerenzanensis
Biotechnology Letters (2023)