Abstract
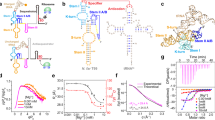
Guanine-responsive riboswitches undergo ligand-dependent structural rearrangements to control gene expression by transcription termination. While the molecular basis for ligand recognition is well established, the associated structural rearrangements and the kinetics involved in the formation of the aptamer domain are less well understood. Using high-resolution optical tweezers, we followed the folding trajectories of a single molecule of the xpt–pbuX guanine aptamer from Bacillus subtilis. We report a rapid six-state conformational rearrangement, in which three of the states are guanine dependent, during the transition from the linear to the native receptor conformation. The folding completes in <1 s. The force-dependent equilibrium kinetics and the mutational data indicated that the flexible J2–J3 junction undergoes a ligand-dependent conformational switching, which triggers the formation of the long-range tertiary interactions and the P1 helix. In the absence of the right ligand, the junction failed to initiate the series of conformational rearrangements required for the riboswitch activities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C. & Breaker, R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003).
Mandal, M. & Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 11, 29–35 (2004).
Batey, R.T., Gilbert, S.D. & Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415 (2004).
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004).
Noeske, J. et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. USA 102, 1372–1377 (2005).
Batey, R.T. Structure and mechanism of purine-binding riboswitches. Q. Rev. Biophys. 45, 345–381 (2012).
Serganov, A. & Patel, D.J. Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol. 22, 279–286 (2012).
Buck, J., Fürtig, B., Noeske, J., Wöhnert, J. & Schwalbe, H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. USA 104, 15699–15704 (2007).
Lee, M.-K., Gal, M., Frydman, L. & Varani, G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. USA 107, 9192–9197 (2010).
Lemay, J.-F., Penedo, J.C., Tremblay, R., Lilley, D.M.J. & Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006).
Rieder, R., Lang, K., Graber, D. & Micura, R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem 8, 896–902 (2007).
Brenner, M.D., Scanlan, M.S., Nahas, M.K., Ha, T. & Silverman, S.K. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry 49, 1596–1605 (2010).
St-Pierre, P., McCluskey, K., Shaw, E., Penedo, J.C. & Lafontaine, D.A. Fluorescence tools to investigate riboswitch structural dynamics. Biochim. Biophys. Acta 1839, 1005–1019 (2014).
Greenleaf, W.J., Frieda, K.L., Foster, D.A.N., Woodside, M.T. & Block, S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633 (2008).
Frieda, K.L. & Block, S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Neupane, K., Yu, H., Foster, D.A., Wang, F. & Woodside, M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 39, 7677–7687 (2011).
Savinov, A., Perez, C.F. & Block, S.M. Single-molecule studies of riboswitch folding. Biochim. Biophys. Acta 1839, 1030–1045 (2014).
Smith, S.B., Cui, Y. & Bustamante, C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 361, 134–162 (2003).
Buck, J., Noeske, J., Wöhnert, J. & Schwalbe, H. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res. 38, 4143–4153 (2010).
Ottink, O.M. et al. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA 13, 2202–2212 (2007).
Liphardt, J., Onoa, B., Smith, S.B., Tinoco, I. Jr. & Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. Science 292, 733–737 (2001).
Onoa, B. et al. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science 299, 1892–1895 (2003).
Noeske, J., Schwalbe, H. & Wöhnert, J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 35, 5262–5273 (2007).
Allnér, O., Nilsson, L. & Villa, A. Loop-loop interaction in an adenine-sensing riboswitch: a molecular dynamics study. RNA 19, 916–926 (2013).
Stoddard, C.D., Gilbert, S.D. & Batey, R.T. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14, 675–684 (2008).
Martick, M. & Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006).
Neuman, K.C., Abbondanzieri, E.A., Landick, R., Gelles, J. & Block, S.M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell 115, 437–447 (2003).
Forde, N.R., Izhaky, D., Woodcock, G.R., Wuite, G.J. & Bustamante, C. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 99, 11682–11687 (2002).
Tinoco, I. Jr. & Bustamante, C. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem. 101-102, 513–533 (2002).
Bell, G.I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978).
Evans, E. & Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 (1997).
Berg-Sorensen, K. & Flyvjerg, H. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 75, 594 (2004).
Smith, S.B., Cui, Y. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Wang, M.D., Yin, H., Landick, R., Gelles, J. & Block, S.M. Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346 (1997).
Wen, J.D. et al. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys. J. 92, 2996–3009 (2007).
Manosas, M. et al. Force unfolding kinetics of RNA using optical tweezers. II. Modeling experiments. Biophys. J. 92, 3010–3021 (2007).
Žoldák, G., Stigler, J., Pelz, B., Li, H. & Rief, M. Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy. Proc. Natl. Acad. Sci. USA 110, 18156–18161 (2013).
Neupane, K. et al. Direct observation of transition paths during the folding of proteins and nucleic acids. Science 352, 239–242 (2016).
Acknowledgements
We would like to thank S. Kumar for technical assistance with the RNA preparation for ITC experiments and B. Plaut for writing Matlab codes. We gratefully acknowledge. S. Smith for helping us with the mini-tweezers. We also acknowledge the Protein Data Bank for letting us use the 3D structure of xpt–pbuX guanine riboswitch aptamer (ID: 4FE5) for illustration purpose. This research was generously supported by NSF CAREER (CHE-1151815) awarded to M.M. and support from Single-Molecule and RNA Biology Institute.
Author information
Authors and Affiliations
Contributions
V.C., H.X. and M.M. collected data in optical tweezers; Z.H. performed the ITC experiments; V.C., H.X., Z.H. and M.M. analyzed data; M.M. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–2 and Supplementary Figures 1–6. (PDF 1129 kb)
Rights and permissions
About this article
Cite this article
Chandra, V., Hannan, Z., Xu, H. et al. Single-molecule analysis reveals multi-state folding of a guanine riboswitch. Nat Chem Biol 13, 194–201 (2017). https://doi.org/10.1038/nchembio.2252
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2252
This article is cited by
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Riboswitch-mediated regulation of riboflavin biosynthesis genes in prokaryotes
3 Biotech (2022)
-
Molecular mechanisms underlying the extreme mechanical anisotropy of the flaviviral exoribonuclease-resistant RNAs (xrRNAs)
Nature Communications (2020)