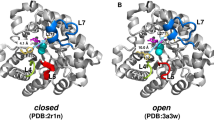
Abstract
Enzymes must be ordered to allow the stabilization of transition states by their active sites, yet dynamic enough to adopt alternative conformations suited to other steps in their catalytic cycles. The biophysical principles that determine how specific protein dynamics evolve and how remote mutations affect catalytic activity are poorly understood. Here we examine a 'molecular fossil record' that was recently obtained during the laboratory evolution of a phosphotriesterase from Pseudomonas diminuta to an arylesterase. Analysis of the structures and dynamics of nine protein variants along this trajectory, and three rationally designed variants, reveals cycles of structural destabilization and repair, evolutionary pressure to 'freeze out' unproductive motions and sampling of distinct conformations with specific catalytic properties in bi-functional intermediates. This work establishes that changes to the conformational landscapes of proteins are an essential aspect of molecular evolution and that change in function can be achieved through enrichment of preexisting conformational sub-states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kirby, A.J. & Hollfelder, F. From Enzyme Models to Model Enzymes (Royal Society of Chemistry, 2009).
Kraut, D.A., Carroll, K.S. & Herschlag, D. Challenges in enzyme mechanism and energetics. Annu. Rev. Biochem. 72, 517–571 (2003).
Ma, B. & Nussinov, R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 14, 652–659 (2010).
Warshel, A. Computer simulations of enzyme catalysis: methods, progress, and insights. Annu. Rev. Biophys. Biomol. Struct. 32, 425–443 (2003).
Elias, M., Wieczorek, G., Rosenne, S. & Tawfik, D.S. The universality of enzymatic rate-temperature dependency. Trends Biochem. Sci. 39, 1–7 (2014).
Bhabha, G. et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 332, 234–238 (2011).
Silva, R.G., Murkin, A.S. & Schramm, V.L. Femtosecond dynamics coupled to chemical barrier crossing in a Born-Oppenheimer enzyme. Proc. Natl. Acad. Sci. USA 108, 18661–18665 (2011).
Bhabha, G. et al. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat. Struct. Mol. Biol. 20, 1243–1249 (2013).
Fraser, J.S. et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 (2009).
Gobeil, S.M. et al. Maintenance of native-like protein dynamics may not be required for engineering functional proteins. Chem. Biol. 21, 1330–1340 (2014).
Jackson, C.J. et al. Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase. Proc. Natl. Acad. Sci. USA 106, 21631–21636 (2009).
Glowacki, D.R., Harvey, J.N. & Mulholland, A.J. Taking Ockham's razor to enzyme dynamics and catalysis. Nat. Chem. 4, 169–176 (2012).
Kamerlin, S.C. & Warshel, A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins 78, 1339–1375 (2010).
O'Brien, P.J. & Hollfelder, F. Hitting a moving target?–Understanding how conformational diversity impacts enzymatic catalysis. Curr. Opin. Chem. Biol. 14, 634–635 (2010).
Fraser, J.S. & Jackson, C.J. Mining electron density for functionally relevant protein polysterism in crystal structures. Cell. Mol. Life Sci. 68, 1829–1841 (2011).
Ramanathan, A., Savol, A., Burger, V., Chennubhotla, C.S. & Agarwal, P.K. Protein conformational populations and functionally relevant substates. Acc. Chem. Res. 47, 149–156 (2014).
Klinman, J.P. & Kohen, A. Evolutionary aspects of enzyme dynamics. J. Biol. Chem. 289, 30205–30212 (2014).
Colletier, J.P. et al. Sampling the conformational energy landscape of a hyperthermophilic protein by engineering key substitutions. Mol. Biol. Evol. 29, 1683–1694 (2012).
Tokuriki, N. & Tawfik, D.S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
James, L.C. & Tawfik, D.S. Conformational diversity and protein evolution–a 60-year-old hypothesis revisited. Trends Biochem. Sci. 28, 361–368 (2003).
Tokuriki, N. et al. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 3, 1257 (2012).
Dellus-Gur, E. et al. Negative epistasis and evolvability in TEM-1 β-lactamase—the thin line between an enzyme's conformational freedom and disorder. J. Mol. Biol. 427, 2396–2409 (2015).
Caldwell, S.R., Newcomb, J.R., Schlecht, K.A. & Raushel, F.M. Limits of diffusion in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry 30, 7438–7444 (1991).
Kaltenbach, M., Jackson, C.J., Campbell, E.C., Hollfelder, F. & Tokuriki, N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. eLife 4, e06492 (2015).
Bora, R.P., Mills, M.J., Frushicheva, M.P. & Warshel, A. On the challenge of exploring the evolutionary trajectory from phosphotriesterase to arylesterase using computer simulations. J. Phys. Chem. B 119, 3434–3445 (2015).
Doncheva, N.T., Assenov, Y., Domingues, F.S. & Albrecht, M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670–685 (2012).
Fetics, S.K. et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015).
Sethi, A., Eargle, J., Black, A.A. & Luthey-Schulten, Z. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. USA 106, 6620–6625 (2009).
Jackson, C.J. et al. In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase. J. Mol. Biol. 375, 1189–1196 (2008).
van den Bedem, H., Bhabha, G., Yang, K., Wright, P.E. & Fraser, J.S. Automated identification of functional dynamic contact networks from X-ray crystallography. Nat. Methods 10, 896–902 (2013).
van den Bedem, H., Dhanik, A., Latombe, J.C. & Deacon, A.M. Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Crystallogr. D Biol. Crystallogr. 65, 1107–1117 (2009).
Burnley, B.T., Afonine, P.V., Adams, P.D. & Gros, P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife 1, e00311 (2012).
Ortlund, E.A., Bridgham, J.T., Redinbo, M.R. & Thornton, J.W. Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548 (2007).
Chao, F.A. et al. Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat. Chem. Biol. 9, 81–83 (2013).
Giger, L. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol. 9, 494–498 (2013).
Khersonsky, O. et al. Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution. J. Mol. Biol. 407, 391–412 (2011).
Preiswerk, N. et al. Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase. Proc. Natl. Acad. Sci. USA 111, 8013–8018 (2014).
Sykora, J. et al. Dynamics and hydration explain failed functional transformation in dehalogenase design. Nat. Chem. Biol. 10, 428–430 (2014).
Morley, K.L. & Kazlauskas, R.J. Improving enzyme properties: when are closer mutations better? Trends Biotechnol. 23, 231–237 (2005).
Love, C.A., Lilley, P.E. & Dixon, N.E. Stable high-copy-number bacteriophage lambda promoter vectors for overproduction of proteins in Escherichia coli. Gene 176, 49–53 (1996).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative, C.P. Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Karplus, P.A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Vagin, A.A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 4, 435–447 (2008).
Schmid, N. et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 (2011).
Berendsen, H., Postma, J., van Gunsteren, W.F. & Hermans, J. in Intermolecular Forces Vol. 14 (ed. Pullman, B.) 331–342 (Springer Netherlands, 1981).
Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A. & Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684 (1984).
Tironi, I.G., Sperb, R., Smith, P.E. & van Gunsteren, W.F. A generalized reaction field method for molecular dynamics simulations. J. Chem. Phys. 102, 5451 (1995).
Heinz, T.N., van Gunsteren, W.F. & Hünenberger, P.H. Comparison of four methods to compute the dielectric permittivity of liquids from molecular dynamics simulations. J. Chem. Phys. 115, 1125 (2001).
Hess, B., Bekker, H., Berendsen, H.J.C. & Fraaije, J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Chem. Phys. 18, 1463–1472 (1997).
Miyamoto, S. & Kollman, P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Bakan, A., Meireles, L.M. & Bahar, I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577 (2011).
Grant, B.J., Rodrigues, A.P., ElSawy, K.M., McCammon, J.A. & Caves, L.S. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696 (2006).
Van Wart, A.T., Durrant, J., Votapka, L. & Amaro, R.E. Weighted Implementation of Suboptimal Paths (WISP): An Optimized Algorithm and Tool for Dynamical Network Analysis. J. Chem. Theory Comput. 10, 511–517 (2014).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 (1996).
Suhre, K. & Sanejouand, Y.H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, W610–W614 (2004).
Acknowledgements
We thank D.S. Tawfik for stimulating discussions. C.J.J. thanks the Australian Research Council for a Future Fellowship (FT140101059) and Discovery Project (DP130102144). This research was undertaken on the MX1 and MX2 beamlines at the Australian Synchrotron, Victoria, Australia. F.H. thanks the Biotechnology and Biological Sciences Research Council and European Research Council (starting investigator grants). M.K. thanks the EU Innovative Training Network (ProSA) for a studentship. N.T. is funded as a Canadian Institutes of Health Research new investigator and a Michael Smith Foundation of Health Research (MSFHR) career investigator. N.T. thanks Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN 418262-12. A.M.B. is funded as a National Health and Medical Research Senior Research Fellow (1022688). This work was supported by the Victorian Life Sciences Computation Initiative, an initiative of the Victorian Government, Australia.
Author information
Authors and Affiliations
Contributions
E.C., M.K., G.J.C., P.D.C., B.T.P., E.K.L. and L.A.-J. performed experiments and analyzed results; A.M.B. and M.W. analyzed results; F.H. conceived the project, designed experiments and analyzed results; N.T. conceived the project, designed experiments, analyzed results and wrote the manuscript; and C.J.J. conceived the project, designed experiments, performed experiments, analyzed results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–7 and Supplementary Tables 1–3. (PDF 1751 kb)
Rights and permissions
About this article
Cite this article
Campbell, E., Kaltenbach, M., Correy, G. et al. The role of protein dynamics in the evolution of new enzyme function. Nat Chem Biol 12, 944–950 (2016). https://doi.org/10.1038/nchembio.2175
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2175
This article is cited by
-
Remote loop evolution reveals a complex biological function for chitinase enzymes beyond the active site
Nature Communications (2024)
-
Frustration can Limit the Adaptation of Promiscuous Enzymes Through Gene Duplication and Specialisation
Journal of Molecular Evolution (2024)
-
Computational remodeling of an enzyme conformational landscape for altered substrate selectivity
Nature Communications (2023)
-
Pervasive epistasis exposes intramolecular networks in adaptive enzyme evolution
Nature Communications (2023)
-
Ratcheting synthesis
Nature Reviews Chemistry (2023)