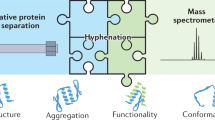
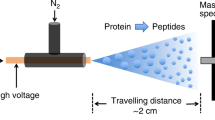
Abstract
To enable the detection of protein conformational isomers, their enzymatic activity and their inhibition in a single experiment, we developed a method based on kinetic capillary electrophoresis coupled on-line with UV detection and ion mobility mass spectrometry (CE–UV–IM–MS). Kinetic CE–UV separated protein conformers and monitored their interconversion dynamics in solution. Ion mobility mass spectrometry analyzed the conformer sizes, exact molecular weights, and structures of an enzyme and of its substrates, inhibitors and corresponding products. This coupled CE–UV–IM–MS system allowed the simultaneous, real-time observation of the effect of small-molecule inhibitors on both the conformational distribution and enzymatic activity of the human tissue transglutaminase TG2. By expanding mass spectrometry profiling of enzymatic reactions beyond proteins and substrates to include protein dynamics, CE–UV–IM–MS opens a new avenue for the modulation and regulation of cellular functions, drug development and protein engineering.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tzeng, S.R. & Kalodimos, C.G. Protein activity regulation by conformational entropy. Nature 488, 236–240 (2012).
Goodey, N.M. & Benkovic, S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 4, 474–482 (2008).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Bruning, J.B. et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 6, 837–843 (2010).
Ruschak, A.M. & Kay, L.E. Proteasome allostery as a population shift between interchanging conformers. Proc. Natl. Acad. Sci. USA 109, E3454–E3462 (2012).
Karplus, M. & Kuriyan, J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA 102, 6679–6685 (2005).
Kar, G., Keskin, O., Gursoy, A. & Nussinov, R. Allostery and population shift in drug discovery. Curr. Opin. Pharmacol. 10, 715–722 (2010).
Nussinov, R. & Tsai, C.J. Allostery in disease and in drug discovery. Cell 153, 293–305 (2013).
Nussinov, R. & Tsai, C.J. The design of covalent allosteric drugs. Annu. Rev. Pharmacol. Toxicol. 55, 249–267 (2015).
Garman, E. 'Cool' crystals: macromolecular cryocrystallography and radiation damage. Curr. Opin. Struct. Biol. 13, 545–551 (2003).
Fraser, J.S. et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA 108, 16247–16252 (2011).
Keedy, D.A. et al. Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR. Structure 22, 899–910 (2014).
Boehr, D.D., Dyson, H.J. & Wright, P.E. An NMR perspective on enzyme dynamics. Chem. Rev. 106, 3055–3079 (2006).
Baldwin, A.J. & Kay, L.E. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 5, 808–814 (2009).
Mulder, F.A., Mittermaier, A., Hon, B., Dahlquist, F.W. & Kay, L.E. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 8, 932–935 (2001).
Tang, C., Schwieters, C.D. & Clore, G.M. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 449, 1078–1082 (2007).
Eisenmesser, E.Z., Bosco, D.A., Akke, M. & Kern, D. Enzyme dynamics during catalysis. Science 295, 1520–1523 (2002).
Watt, E.D., Shimada, H., Kovrigin, E.L. & Loria, J.P. The mechanism of rate-limiting motions in enzyme function. Proc. Natl. Acad. Sci. USA 104, 11981–11986 (2007).
Kempf, J.G., Jung, J.Y., Ragain, C., Sampson, N.S. & Loria, J.P. Dynamic requirements for a functional protein hinge. J. Mol. Biol. 368, 131–149 (2007).
Korzhnev, D.M. et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430, 586–590 (2004).
Tang, Y., Grey, M.J., McKnight, J., Palmer, A.G. III & Raleigh, D.P. Multistate folding of the villin headpiece domain. J. Mol. Biol. 355, 1066–1077 (2006).
Korzhnev, D.M. & Kay, L.E. Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res. 41, 442–451 (2008).
Smith, M.J. et al. Real-time NMR monitoring of biological activities in complex physiological environments. Curr. Opin. Struct. Biol. 32, 39–47 (2015).
Bernadó, P., Mylonas, E., Petoukhov, M.V., Blackledge, M. & Svergun, D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007).
Fischetti, R.F., Rodi, D.J., Gore, D.B. & Makowski, L. Wide-angle X-ray solution scattering as a probe of ligand-induced conformational changes in proteins. Chem. Biol. 11, 1431–1443 (2004).
Nguyen, H.T., Pabit, S.A., Meisburger, S.P., Pollack, L. & Case, D.A. Accurate small and wide angle x-ray scattering profiles from atomic models of proteins and nucleic acids. J. Chem. Phys. 141, 22D508 (2014).
Heyduk, T. Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 13, 292–296 (2002).
Truong, K. & Ikura, M. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 11, 573–578 (2001).
Shi, H., Pierson, N.A., Valentine, S.J. & Clemmer, D.E. Conformation types of ubiquitin [M+8H]8+ ions from water:methanol solutions: evidence for the N and A States in aqueous solution. J. Phys. Chem. B 116, 3344–3352 (2012).
Smith, D.P., Giles, K., Bateman, R.H., Radford, S.E. & Ashcroft, A.E. Monitoring copopulated conformational states during protein folding events using electrospray ionization-ion mobility spectrometry-mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 2180–2190 (2007).
Nabuchi, Y., Hirose, K. & Takayama, M. Ion mobility and collision-induced dissociation analysis of carbonic anhydrase 2. Anal. Chem. 82, 8890–8896 (2010).
Konijnenberg, A., Butterer, A. & Sobott, F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta 1834, 1239–1256 (2013).
Kaddis, C.S. & Loo, J.A. Native protein MS and ion mobility large flying proteins with ESI. Anal. Chem. 79, 1778–1784 (2007).
Wolynes, P.G. Biomolecular folding in vacuo!!!(?). Proc. Natl. Acad. Sci. USA 92, 2426–2427 (1995).
Jurneczko, E. & Barran, P.E. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst. 136, 20–28 (2011).
Berezovski, M. & Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures—a single experiment reveals equilibrium and kinetic parameters of protein-DNA interactions. J. Am. Chem. Soc. 124, 13674–13675 (2002).
Heegaard, N.H., Rovatti, L., Nissen, M.H. & Hamdan, M. Structural and conformational variants of human beta2-microglobulin characterized by capillary electrophoresis and complementary separation methods. J. Chromatogr. A 1004, 51–59 (2003).
Mironov, G.G. et al. Bioanalysis for biocatalysis: multiplexed capillary electrophoresis-mass spectrometry assay for aminotransferase substrate discovery and specificity profiling. J. Am. Chem. Soc. 135, 13728–13736 (2013).
Liu, S., Cerione, R.A. & Clardy, J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. USA 99, 2743–2747 (2002).
Clouthier, C.M., Mironov, G.G., Okhonin, V., Berezovski, M.V. & Keillor, J.W. Real-time monitoring of protein conformational dynamics in solution using kinetic capillary electrophoresis. Angew. Chem. Int. Edn Engl. 51, 12464–12468 (2012).
Gnaccarini, C., Ben-Tahar, W., Lubell, W.D., Pelletier, J.N. & Keillor, J.W. Fluorometric assay for tissue transglutaminase-mediated transamidation activity. Bioorg. Med. Chem. 17, 6354–6359 (2009).
Hu, B.H. & Messersmith, P.B. Rational design of transglutaminase substrate peptides for rapid enzymatic formation of hydrogels. J. Am. Chem. Soc. 125, 14298–14299 (2003).
Caron, N.S., Munsie, L.N., Keillor, J.W. & Truant, R. Using FLIM-FRET to measure conformational changes of transglutaminase type 2 in live cells. PLoS One 7, e44159 (2012).
Keillor, J.W. et al. The bioorganic chemistry of transglutaminase — from mechanism to inhibition and engineering. Can. J. Chem. 86, 271–276 (2008).
Grant, B.J., Gorfe, A.A. & McCammon, J.A. Large conformational changes in proteins: signaling and other functions. Curr. Opin. Struct. Biol. 20, 142–147 (2010).
Mattevi, A., Rizzi, M. & Bolognesi, M. New structures of allosteric proteins revealing remarkable conformational changes. Curr. Opin. Struct. Biol. 6, 824–829 (1996).
Lipscomb, W.N. Multisubunit allosteric proteins. in Protein Dynamics, Function, and Design (eds. Jardetzky, O., Lefèvre, J.-F. &. Holbrook, R.E.) 27–35 (Springer, 1998).
Pinkas, D.M., Strop, P., Brunger, A.T. & Khosla, C. Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 5, e327 (2007).
Roy, I., Smith, O., Clouthier, C.M. & Keillor, J.W. Expression, purification and kinetic characterisation of human tissue transglutaminase. Protein Expr. Purif. 87, 41–46 (2013).
Lubell, W.D., Blankenship, J.W., Fridkin, G. & Kaul, R. Peptides: Science of Synthesis (Thieme, Stuttgart, 2005).
Leblanc, A., Gravel, C., Labelle, J. & Keillor, J.W. Kinetic studies of guinea pig liver transglutaminase reveal a general-base-catalyzed deacylation mechanism. Biochemistry 40, 8335–8342 (2001).
Stone, S.R. & Hofsteenge, J. Specificity of activated human protein C. Biochem. J. 230, 497–502 (1985).
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (M.V.B. and J.W.K.), the Canada Foundation for Innovation (M.V.B.), the Ministry of Research and Innovation, Ontario, Canada (M.V.B.) and by an Ontario Graduate Scholarship (for G.G.M.).
Author information
Authors and Affiliations
Contributions
G.G.M., J.W.K., and M.V.B. conceived of the project, designed the experiments, analyzed the data, and wrote the manuscript. G.G.M. built the CE–UV–IM–MS system and performed all CE–UV, IM–MS and CE–UV–IM–MS experiments, conformational analysis, and inhibition analysis. C.M.C. expressed and purified TG2 protein, synthesized peptide substrates, and assisted in writing the manuscript; A.A. synthesized inhibitors, measured inhibition kinetics with a chromogenic substrate, and assisted in writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–3 and Supplementary Table 1. (PDF 374 kb)
Supplementary Note
Supplementary Note (PDF 1201 kb)
Rights and permissions
About this article
Cite this article
Mironov, G., Clouthier, C., Akbar, A. et al. Simultaneous analysis of enzyme structure and activity by kinetic capillary electrophoresis–MS. Nat Chem Biol 12, 918–922 (2016). https://doi.org/10.1038/nchembio.2170
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2170
This article is cited by
-
Studying protein structure and function by native separation–mass spectrometry
Nature Reviews Chemistry (2022)
-
Sulfated glycosaminoglycans inhibit transglutaminase 2 by stabilizing its closed conformation
Scientific Reports (2022)
-
Capillary zone electrophoresis coupled to drift tube ion mobility-mass spectrometry for the analysis of native and APTS-labeled N-glycans
Analytical and Bioanalytical Chemistry (2019)