Abstract
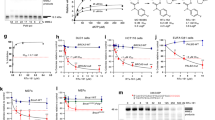
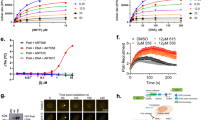
CD437 is a retinoid-like small molecule that selectively induces apoptosis in cancer cells, but not in normal cells, through an unknown mechanism. We used a forward-genetic strategy to discover mutations in POLA1 that coincide with CD437 resistance (POLA1R). Introduction of one of these mutations into cancer cells by CRISPR-Cas9 genome editing conferred CD437 resistance, demonstrating causality. POLA1 encodes DNA polymerase α, the enzyme responsible for initiating DNA synthesis during the S phase of the cell cycle. CD437 inhibits DNA replication in cells and recombinant POLA1 activity in vitro. Both effects are abrogated by the identified POLA1 mutations, supporting POLA1 as the direct antitumor target of CD437. In addition, we detected an increase in the total fluorescence intensity and anisotropy of CD437 in the presence of increasing concentrations of POLA1 that is consistent with a direct binding interaction. The discovery of POLA1 as the direct anticancer target for CD437 has the potential to catalyze the development of CD437 into an anticancer therapeutic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Holmes, W.F., Soprano, D.R. & Soprano, K.J. Comparison of the mechanism of induction of apoptosis in ovarian carcinoma cells by the conformationally restricted synthetic retinoids CD437 and 4-HPR. J. Cell. Biochem. 89, 262–278 (2003).
Hsu, S.L., Yin, S.C., Liu, M.C., Reichert, U. & Ho, W.L. Involvement of cyclin-dependent kinase activities in CD437-induced apoptosis. Exp. Cell Res. 252, 332–341 (1999).
Schadendorf, D. et al. Treatment of melanoma cells with the synthetic retinoid CD437 induces apoptosis via activation of AP-1 in vitro, and causes growth inhibition in xenografts in vivo. J. Cell Biol. 135, 1889–1898 (1996).
Sun, S.Y., Yue, P., Shroot, B., Hong, W.K. & Lotan, R. Induction of apoptosis in human non-small cell lung carcinoma cells by the novel synthetic retinoid CD437. J. Cell. Physiol. 173, 279–284 (1997).
Rees, M.G. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12, 109–116 (2016).
Lotan, R. Receptor-independent induction of apoptosis by synthetic retinoids. J. Biol. Regul. Homeost. Agents 17, 13–28 (2003).
Pfahl, M. & Piedrafita, F.J. Retinoid targets for apoptosis induction. Oncogene 22, 9058–9062 (2003).
Rishi, A.K. et al. Post-transcriptional regulation of the DNA damage-inducible gadd45 gene in human breast carcinoma cells exposed to a novel retinoid CD437. Nucleic Acids Res. 27, 3111–3119 (1999).
Hail, N. Jr. & Lotan, R. Synthetic retinoid CD437 promotes rapid apoptosis in malignant human epidermal keratinocytes and G1 arrest in their normal counterparts. J. Cell. Physiol. 186, 24–34 (2001).
Cincinelli, R. et al. A novel atypical retinoid endowed with proapoptotic and antitumor activity. J. Med. Chem. 46, 909–912 (2003).
Cincinelli, R. et al. Synthesis and structure-activity relationships of a new series of retinoid-related biphenyl-4-ylacrylic acids endowed with antiproliferative and proapoptotic activity. J. Med. Chem. 48, 4931–4946 (2005).
Martin, B. et al. Selective synthetic ligands for human nuclear retinoic acid receptors. Skin Pharmacol. 5, 57–65 (1992).
Sun, S.Y. et al. Identification of receptor-selective retinoids that are potent inhibitors of the growth of human head and neck squamous cell carcinoma cells. Clin. Cancer Res. 6, 1563–1573 (2000).
Sun, S.Y. et al. Dual mechanisms of action of the retinoid CD437: nuclear retinoic acid receptor-mediated suppression of squamous differentiation and receptor-independent induction of apoptosis in UMSCC22B human head and neck squamous cell carcinoma cells. Mol. Pharmacol. 58, 508–514 (2000).
Hsu, C.A. et al. Retinoid induced apoptosis in leukemia cells through a retinoic acid nuclear receptor-independent pathway. Blood 89, 4470–4479 (1997).
Parrella, E. et al. Antitumor activity of the retinoid-related molecules (E)-3-(4′-hydroxy-3′-adamantylbiphenyl-4-yl)acrylic acid (ST1926) and 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid (CD437) in F9 teratocarcinoma: role of retinoic acid receptor gamma and retinoid-independent pathways. Mol. Pharmacol. 70, 909–924 (2006).
Wacker, S.A., Houghtaling, B.R., Elemento, O. & Kapoor, T.M. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 8, 235–237 (2012).
Glaab, W.E. & Tindall, K.R. Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis 18, 1–8 (1997).
Baranovskiy, A.G. et al. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 (2014).
Mishur, R.J., Griffin, M.E., Battle, C.H., Shan, B. & Jayawickramarajah, J. Molecular recognition and enhancement of aqueous solubility and bioactivity of CD437 by β-cyclodextrin. Bioorg. Med. Chem. Lett. 21, 857–860 (2011).
Ciccia, A. & Elledge, S.J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Valli, C. et al. Atypical retinoids ST1926 and CD437 are S-phase-specific agents causing DNA double-strand breaks: significance for the cytotoxic and antiproliferative activity. Mol. Cancer Ther. 7, 2941–2954 (2008).
Finn, R.S. et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 16, 25–35 (2015).
Ikegami, S. et al. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275, 458–460 (1978).
Sessa, C. et al. Phase I and clinical pharmacological evaluation of aphidicolin glycinate. J. Natl. Cancer Inst. 83, 1160–1164 (1991).
Edelson, R.E., Gorycki, P.D. & MacDonald, T.L. The mechanism of aphidicolin bioinactivation by rat liver in vitro systems. Xenobiotica 20, 273–287 (1990).
Prasad, G., Edelson, R.A., Gorycki, P.D. & Macdonald, T.L. Structure-activity relationships for the inhibition of DNA polymerase alpha by aphidicolin derivatives. Nucleic Acids Res. 17, 6339–6348 (1989).
Sun, S.Y. et al. Mechanisms of apoptosis induced by the synthetic retinoid CD437 in human non-small cell lung carcinoma cells. Oncogene 18, 2357–2365 (1999).
Pedrali-Noy, G. et al. Synchronization of HeLa cell cultures by inhibition of DNA polymerase alpha with aphidicolin. Nucleic Acids Res. 8, 377–387 (1980).
Patel, R.K. & Jain, M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7, e30619 (2012).
Hashiguchi, K. et al. Involvement of ETS1 in thioredoxin-binding protein 2 transcription induced by a synthetic retinoid CD437 in human osteosarcoma cells. Biochem. Biophys. Res. Commun. 391, 621–626 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Pruitt, K.D. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 (2014).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Acknowledgements
The authors thank J. Ready, S.L. McKnight, and members of the Nijhawan laboratory for helpful comments. We thank the McDermott Sequencing Center at UT Southwestern Medical Center for Illumina sequencing, T. Tahirov for the pFastBac1 POLA1 plasmid, J. McKnight for site-directed mutagenesis of POLA1, and B. Li, H. Yu and L. Beatty for help with Sf9 culture. T.H. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. S.B.P. was supported by a grant to M.K. Rosen from NIGMS (R01-GM56322). J.K. was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP150596. This research was supported by a Harold C. Simmons Cancer Center Startup Awards, a Disease Oriented Clinical Scholar (DOCS) award, a Damon Runyon Clinical Investigator award (CI-68-13) and a grant from the Welch Foundation (I-1879) to D.N.
Author information
Authors and Affiliations
Contributions
T.H. designed the study, performed most of the experiments, interpreted the results, and wrote the manuscript. M.G. performed the initial screen and contributed to validation studies. E.C. expressed and purified recombinant POLA1 protein. S.B.P. guided the binding assay and analyzed the binding data. J.K. and Y.X. performed all bioinformatics analyses. D.N. designed and supervised the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–7. (PDF 18963 kb)
Supplementary Data Set 1
Mutation results for exome sequencing of HCT-116 clones. A list of all detected non-synonymous, splice site, and insertion/deletion mutations for 19 HCT-116 clones. 6 of these clones were resistant to CD437 (CD437R) and 13 others were sensitive (Control). (XLSX 3448 kb)
Supplementary Data Set 2
Genes with missense mutations in HCT-116 clones. A list of genes for which exome sequencing revealed missense mutations in at least one of the 19 HCT-116 clones. Genes that harbored missense mutations in CD437R clones but not Control clones are highlighted and listed at the top. (XLSX 506 kb)
Rights and permissions
About this article
Cite this article
Han, T., Goralski, M., Capota, E. et al. The antitumor toxin CD437 is a direct inhibitor of DNA polymerase α. Nat Chem Biol 12, 511–515 (2016). https://doi.org/10.1038/nchembio.2082
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2082
This article is cited by
-
Replication-associated formation and repair of human topoisomerase IIIα cleavage complexes
Nature Communications (2023)
-
The splicing factor SNRPB promotes ovarian cancer progression through regulating aberrant exon skipping of POLA1 and BRCA2
Oncogene (2023)
-
Targeting pan-essential pathways in cancer with cytotoxic chemotherapy: challenges and opportunities
Cancer Chemotherapy and Pharmacology (2023)
-
Synthesis and in vitro antibacterial, antifungal, anti-proliferative activities of novel adamantane-containing thiazole compounds
Scientific Reports (2022)
-
MIF is a 3’ flap nuclease that facilitates DNA replication and promotes tumor growth
Nature Communications (2021)