Abstract
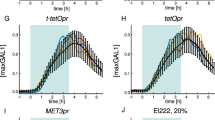
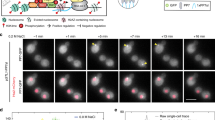
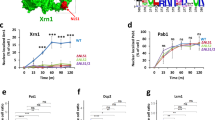
We engineered a photoactivatable system for rapidly and reversibly exporting proteins from the nucleus by embedding a nuclear export signal in the LOV2 domain from phototropin 1. Fusing the chromatin modifier Bre1 to the photoswitch, we achieved light-dependent control of histone H2B monoubiquitylation in yeast, revealing fast turnover of the ubiquitin mark. Moreover, this inducible system allowed us to dynamically monitor the status of epigenetic modifications dependent on H2B ubiquitylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tischer, D. & Weiner, O.D. Nat. Rev. Mol. Cell Biol. 15, 551–558 (2014).
Yang, X., Jost, A.P.-T., Weiner, O.D. & Tang, C. Mol. Biol. Cell 24, 2419–2430 (2013).
Gautier, A. et al. J. Am. Chem. Soc. 132, 4086–4088 (2010).
Yumerefendi, H. et al. PLoS One 10, e0128443 (2015).
Niopek, D. et al. Nat. Comm. 5, 4404 (2014).
Harper, S.M., Neil, L.C. & Gardner, K.H. Science 301, 1541–1544 (2003).
Zayner, J.P., Antoniou, C. & Sosnick, T.R. J. Mol. Biol. 419, 61–74 (2012).
Hutten, S. & Kehlenbach, R.H. Trends Cell Biol. 17, 193–201 (2007).
Güttler, T. et al. Nat. Struct. Mol. Biol. 17, 1367–1376 (2010).
Hodel, M.R., Corbett, A.H. & Hodel, A.E. J. Biol. Chem. 276, 1317–1325 (2001).
Guntas, G. et al. Proc. Natl. Acad. Sci. USA 112, 112–117 (2015).
Hodel, A.E. et al. J. Biol. Chem. 281, 23545–23556 (2006).
Xiao, T. et al. Mol. Cell. Biol. 25, 637–651 (2005).
Fuchs, G., Hollander, D., Voichek, Y., Ast, G. & Oren, M. Genome Res. 24, 1572–1583 (2014).
Henry, K.W. et al. Genes Dev. 17, 2648–2663 (2003).
Kosugi, S., Hasebe, M., Tomita, M. & Yanagawa, H. Proc. Natl. Acad. Sci. USA 106, 10171–10176 (2009).
Sun, Z.-W. & Allis, C.D. Nature 418, 104–108 (2002).
Briggs, S.D. et al. Nature 418, 498 (2002).
Huang, F. et al. J. Biol. Chem. http://dx.doi.org/10.1074/jbc.M115.693085 (2015).
Katan-Khaykovich, Y. & Struhl, K. EMBO J. 24, 2138–2149 (2005).
Cai, L., Makhov, A.M., Schafer, D.A. & Bear, J.E. Cell 134, 828–842 (2008).
Hallett, R.A., Zimmerman, S.P., Yumerefendi, H., Bear, J.E. & Kuhlman, B. ACS Synth. Biol. 5, 53–64 (2016).
Gietz, R.D. & Schiestl, R.H. Nat. Protoc. 2, 35–37 (2007).
Acknowledgements
We would like to thank D. Dickinson for help with C. elegans injections, H. Meriesh for providing a plasmid containing BRE1, and R. Dronamraju and Z.-W. Sun for helpful discussions. Funding: J.E.B. (NIH GM111557), B.K., K.H., B.S. (NIH DA036877), K.H. (GM102924).
Author information
Authors and Affiliations
Contributions
H.Y. and B.K. conceived the project and designed the experiments. H.Y., A.M.L. and S.P.Z. performed the experiments. H.Y., A.M.L., S.P.Z., B.D.S. and B.K. analyzed the results. K.H. and J.E.B. provided reagents and equipment. H.Y. and B.K. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–13, Supplementary Tables 1–4 and Supplementary Note. (PDF 19788 kb)
LINXa3 blue light single activation and reversion in mouse fibroblast cells (IA32)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 50 μm. (MOV 6347 kb)
LINXb3 blue light single activation and reversion in mouse fibroblast cells (IA32)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 50 μm. (MOV 8240 kb)
Two cycles of blue light activation and reversion of LINXa3 in mouse fibroblast cells (IA32)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 100 μm. (MOV 9768 kb)
Two cycles of blue light activation and reversion of LINXb3 in mouse fibroblast cells (IA32)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 50 μm. (MOV 22837 kb)
Activation and reversion of LINXa3 in the C. elegans embryo
Activation is performed on the entire field of view and is indicated with words at the upper left corner. Scale bar is 10 μm. (MOV 14408 kb)
Two cycles of blue light activation and reversion of LINXa3-nano (red color) and iLID-Mito (green color) in mouse fibroblast cells (IA32)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 50 μm. (MOV 28412 kb)
Activation and reversion of LINXa4-Bre1 (green color) in an H2B-mCherry (labels the nucleus in red) yeast strain (Supplementary Table 3)
The appearance and disappearance of a square indicate the area and the time of blue light activation. Scale bar is 10 μm. (MOV 9449 kb)
Rights and permissions
About this article
Cite this article
Yumerefendi, H., Lerner, A., Zimmerman, S. et al. Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Nat Chem Biol 12, 399–401 (2016). https://doi.org/10.1038/nchembio.2068
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2068
This article is cited by
-
In vivo imaging of inflammatory response in cancer research
Inflammation and Regeneration (2023)
-
The emergence of molecular systems neuroscience
Molecular Brain (2022)
-
Nanoscale programming of cellular and physiological phenotypes: inorganic meets organic programming
npj Systems Biology and Applications (2021)
-
Optogenetic control of protein binding using light-switchable nanobodies
Nature Communications (2020)
-
Near-infrared light–controlled systems for gene transcription regulation, protein targeting and spectral multiplexing
Nature Protocols (2018)