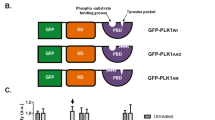
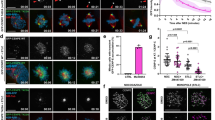
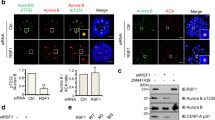
Abstract
Protein kinase signaling along the kinetochore–centromere axis is crucial to assure mitotic fidelity, yet the details of its spatial coordination are obscure. Here, we examined how pools of human Polo-like kinase 1 (Plk1) within this axis control signaling events to elicit mitotic functions. To do this, we restricted active Plk1 to discrete subcompartments within the kinetochore–centromere axis using chemical genetics and decoded functional and phosphoproteomic signatures of each. We observe distinct phosphoproteomic and functional roles, suggesting that Plk1 exists and functions in discrete pools along this axis. Deep within the centromere, Plk1 operates to assure proper chromosome alignment and segregation. Thus, Plk1 at the kinetochore is a conglomerate of an observable bulk pool coupled with additional functional pools below the threshold of microscopic detection or resolution. Although complex, this multiplicity of locales provides an opportunity to decouple functional and phosphoproteomic signatures for a comprehensive understanding of Plk1's kinetochore functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Godek, K.M., Kabeche, L. & Compton, D.A. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 16, 57–64 (2015).
London, N. & Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 736–747 (2014).
Foley, E.A. & Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14, 25–37 (2013).
Liu, D., Vader, G., Vromans, M.J.M., Lampson, M.A. & Lens, S.M.A. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353 (2009).
Welburn, J.P.I. et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38, 383–392 (2010).
Barr, F.A., Silljé, H.H.W. & Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 (2004).
Arnaud, L., Pines, J. & Nigg, E.A. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma 107, 424–429 (1998).
Elia, A.E.H. et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95 (2003).
Elia, A.E.H., Cantley, L.C. & Yaffe, M.B. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–1231 (2003).
Hanisch, A., Wehner, A., Nigg, E.A. & Silljé, H.H.W. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol. Biol. Cell 17, 448–459 (2006).
Liu, D., Davydenko, O. & Lampson, M.A. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J. Cell Biol. 198, 491–499 (2012).
Lera, R.F. & Burkard, M.E. High mitotic activity of Polo-like kinase 1 is required for chromosome segregation and genomic integrity in human epithelial cells. J. Biol. Chem. 287, 42812–42825 (2012).
Park, J.-E., Erikson, R.L. & Lee, K.S. Feed-forward mechanism of converting biochemical cooperativity to mitotic processes at the kinetochore plate. Proc. Natl. Acad. Sci. USA 108, 8200–8205 (2011).
Burkard, M.E. et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7, e1000111 (2009).
Qi, W., Tang, Z. & Yu, H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell 17, 3705–3716 (2006).
Nishino, M. et al. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr. Biol. 16, 1414–1421 (2006).
Suijkerbuijk, S.J.E., Vleugel, M., Teixeira, A. & Kops, G.J.P.L. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell 23, 745–755 (2012).
Elowe, S., Hümmer, S., Uldschmid, A., Li, X. & Nigg, E.A. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219 (2007).
Maia, A.R.R. et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J. Cell Biol. 199, 285–301 (2012).
Hood, E.A., Kettenbach, A.N., Gerber, S.A. & Compton, D.A. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol. Biol. Cell 23, 2264–2274 (2012).
Kang, Y.H. et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell 24, 409–422 (2006).
Park, C.H. et al. Mammalian Polo-like kinase 1 (Plk1) promotes proper chromosome segregation by phosphorylating and delocalizing the PBIP1·CENP-Q complex from kinetochores. J. Biol. Chem. 290, 8569–8581 (2015).
Goto, H. et al. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8, 180–187 (2006).
Chu, Y. et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 3, 260–267 (2011).
Colnaghi, R. & Wheatley, S.P. Liaisons between survivin and Plk1 during cell division and cell death. J. Biol. Chem. 285, 22592–22604 (2010).
Kothe, M. et al. Selectivity-determining residues in Plk1. Chem. Biol. Drug Des. 70, 540–546 (2007).
Cheerambathur, D.K. & Desai, A. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 26, 113–122 (2014).
Oppermann, F.S. et al. Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol. Cell. Proteomics 11, O111.012351 (2012).
Grosstessner-Hain, K. et al. Quantitative phospho-proteomics to investigate the Polo-like kinase 1-dependent phospho-proteome. Mol. Cell. Proteomics 10, M111.008540 (2011).
Kettenbach, A.N. et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 (2011).
Santamaria, A. et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10, M110.004457 (2011).
Burkard, M.E. et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 104, 4383–4388 (2007).
Burkard, M.E., Santamaria, A. & Jallepalli, P.V. Enabling and disabling polo-like kinase 1 inhibition through chemical genetics. ACS Chem. Biol. 7, 978–981 (2012).
Suzuki, A., Badger, B.L., Wan, X., DeLuca, J.G. & Salmon, E.D. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper intrakinetochore stretch. Dev. Cell 30, 717–730 (2014).
Wan, X. et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672–684 (2009).
Domnitz, S.B., Wagenbach, M., Decarreau, J. & Wordeman, L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 197, 231–237 (2012).
Honnappa, S. et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376 (2009).
Phanstiel, D., Unwin, R., McAlister, G.C. & Coon, J.J. Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 1693–1698 (2009).
Maney, T., Hunter, A.W., Wagenbach, M. & Wordeman, L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787–801 (1998).
Carmena, M. et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 10, e1001250 (2012).
Baumann, C., Körner, R., Hofmann, K. & Nigg, E.A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 (2007).
Lowery, D.M. et al. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 26, 2262–2273 (2007).
Kang, Y.H. et al. Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores. J. Biol. Chem. 286, 19744–19757 (2011).
Meppelink, A., Kabeche, L., Vromans, M.J.M., Compton, D.A. & Lens, S.M.A. Shugoshin-1 balances Aurora B kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep. 11, 508–515 (2015).
Foley, E.A., Maldonado, M. & Kapoor, T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13, 1265–1271 (2011).
Liu, D. et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188, 809–820 (2010).
Beck, J. et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 15, 430–439 (2013).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Hornbeck, P.V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Wenger, C.D., Phanstiel, D.H., Lee, M.V., Bailey, D.J. & Coon, J.J. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 (2011).
Taus, T. et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 (2011).
Acknowledgements
This work was supported by NIH R01 GM097245 (to M.E.B.), NIH R01 GM080148 (to J.J.C.), NIH R01 GM024364 (to E.D.S.), Cancer Center Support P30 CA014520, and http://www.Effcansah.com. The authors thank I.M. Cheeseman, S.S. Taylor, and T.J. Yen for contributing reagents, and B.A. Weaver and members of the Burkard laboratory for helpful discussions.
Author information
Authors and Affiliations
Contributions
R.F.L. and M.E.B. designed the research. R.F.L., G.K.P., A.S., and J.M.J. performed experiments. R.F.L., G.K.P., A.S., J.M.J., and M.E.B. analyzed the data. M.E.B., J.J.C., and E.D.S. supervised the research. R.F.L. and M.E.B. drafted the manuscript. All authors revised and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–12 and Supplementary Tables 1–3. (PDF 2968 kb)
Supplementary Data Set 1
Results table listing phosphopeptides encountered by mass spectrometry of Plk1 cell lines labeled with tandem mass tags. A total of 11,407 phosphopeptides were encountered from the 10 cell fractions (5 cell lines A BI- 2536). The raw 10-plex reporter ion intensities of localized phosphopeptide isoforms were log2 transformed and normalized against inhibited Plk1wt (+BI-2536) to obtain the relative phosphopeptide and protein quantitation for each cell line and condition. Terminal columns in the table indicate relative phosphopeptide expression for individual lines after BI-2536 treatment. See Online Methods section for details of analysis. (XLSX 8750 kb)
Rights and permissions
About this article
Cite this article
Lera, R., Potts, G., Suzuki, A. et al. Decoding Polo-like kinase 1 signaling along the kinetochore–centromere axis. Nat Chem Biol 12, 411–418 (2016). https://doi.org/10.1038/nchembio.2060
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2060