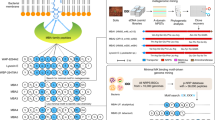
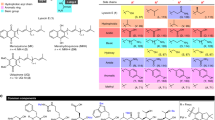
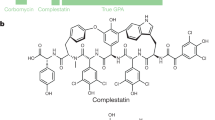
Abstract
Antibiotics are essential for numerous medical procedures, including the treatment of bacterial infections, but their widespread use has led to the accumulation of resistance, prompting calls for the discovery of antibacterial agents with new targets. A majority of clinically approved antibacterial scaffolds are derived from microbial natural products, but these valuable molecules are not well annotated or organized, limiting the efficacy of modern informatic analyses. Here, we provide a comprehensive resource defining the targets, chemical origins and families of the natural antibacterial collective through a retrobiosynthetic algorithm. From this we also detail the directed mining of biosynthetic scaffolds and resistance determinants to reveal structures with a high likelihood of having previously unknown modes of action. Implementing this pipeline led to investigations of the telomycin family of natural products from Streptomyces canus, revealing that these bactericidal molecules possess a new antibacterial mode of action dependent on the bacterial phospholipid cardiolipin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Bush, K. et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896 (2011).
Fischbach, M.A. & Walsh, C.T. Antibiotics for emerging pathogens. Science 325, 1089–1093 (2009).
Newman, D.J. & Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012).
Vining, C.L. Roles of secondary metabolites from microbes. in CIBA Foundation Symposium 171—Secondary Metabolites: Their Function and Evolution 184–198 (Wiley, Chichester, 1992).
Fischbach, M.A. & Clardy, J. One pathway, many products. Nat. Chem. Biol. 3, 353–355 (2007).
Payne, D.J., Gwynn, M.N., Holmes, D.J. & Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Baumann, S. et al. Cystobactamids: myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew. Chem. Int. Edn. Engl. 53, 14605–14609 (2014).
Lin, A.H., Murray, R.W., Vidmar, T.J. & Marotti, K.R. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41, 2127–2131 (1997).
Keller, S., Schadt, H.S., Ortel, I. & Süssmuth, R.D. Action of atrop-abyssomicin C as an inhibitor of 4-amino-4-deoxychorismate synthase PabB. Angew. Chem. Int. Edn. Engl. 46, 8284–8286 (2007).
Li, J.W. & Vederas, J.C. Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165 (2009).
Cundliffe, E. & Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 37, 643–672 (2010).
D'Costa, V.M., McGrann, K.M., Hughes, D.W. & Wright, G.D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Thaker, M.N. et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922–927 (2013).
Bibikova, M.V., Ivanitskaia, L.P. & Singal, E.M. [Directed screening of aminoglycoside antibiotic producers on selective media with gentamycin]. Antibiotiki 26, 488–492 (1981).
Ivanitskaia, L.P., Bibikova, M.V., Gromova, M.N., Zhdanovich, IuV. & Istratov, E.N. [Use of selective media with lincomycin for the directed screening of antibiotic producers]. Antibiotiki 26, 83–86 (1981).
Forsberg, K.J. et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111 (2012).
Boucher, H.W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Doroghazi, J.R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Donadio, S., Maffioli, S., Monciardini, P., Sosio, M. & Jabes, D. Antibiotic discovery in the twenty-first century: current trends and future perspectives. J. Antibiot. 63, 423–430 (2010).
Walsh, C.T. & Wencewicz, T.A. Prospects for new antibiotics: a molecule-centered perspective. J. Antibiot. 67, 7–22 (2014).
Goss, R.J.M., Shankar, S. & Fayad, A.A. The generation of “unnatural” products: synthetic biology meets synthetic chemistry. Nat. Prod. Rep. 29, 870–889 (2012).
Ling, L.L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Cociancich, S. et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat. Chem. Biol. 11, 195–197 (2015).
Hamamoto, H. et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat. Chem. Biol. 11, 127–133 (2015).
Weber, T. et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W1, W237-43 (2015).
Medema, M.H. et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 11, 625–631 (2015).
Hadjithomas, M. et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio. 6, e00932–15 (2015).
Bérdy, J. et al. Handbook of Antibiotic Compounds Vols. I–X (CRC Press, Boca Raton, Florida, USA, 1980–1982).
Bérdy, J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395 (2012).
Koch, M.A. et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP). Proc. Natl. Acad. Sci. USA 102, 17272–17277 (2005).
Over, B. et al. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 5, 21–28 (2013).
Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48 (2014).
Srivastava, A. et al. New target for inhibition of bacterial RNA polymerase: 'switch region'. Curr. Opin. Microbiol. 14, 532–543 (2011).
Skinnider, M.A. et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43, 9645–9662 (2015).
Gibson, M.K., Forsberg, K.J. & Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015).
Nakao, M. et al. Pyloricidins, novel anti-Helicobacter pylori antibiotics produced by Bacillus sp. I. Taxonomy, fermentation and biological activity. J. Antibiot. 54, 926–933 (2001).
Nakajima, M. et al. Mycoplanecins, novel antimycobacterial antibiotics from Actinoplanes awajinensis subsp. mycoplanecinus subsp. nov. II. Isolation, physico-chemical characterization and biological activities of mycoplanecin A. J. Antibiot. 36, 961–966 (1983).
Misiek, M. et al. Telomycin, a new antibiotic. Antibiot. Annu. 5, 852–855 (1957–1958).
Gourevitch, A. et al. Microbiological studies on telomycin. Antibiot. Annu. 5, 856–862 (1957–1958).
Tisch, D.E., Huftalen, J.B. & Dickison, H.L. Pharmacological studies with telomycin. Antibiot. Annu. 5, 863–868 (1957–1958).
Sheehan, J.C., Mania, D., Nakamura, S., Stock, J.A. & Maeda, K. The structure of telomycin. J. Am. Chem. Soc. 90, 462–470 (1968).
Oliva, B., Maiese, W.M., Greenstein, M., Borders, D.B. & Chopra, I. Mode of action of the cyclic depsipeptide antibiotic LL-AO341β1 and partial characterization of a Staphylococcus aureus mutant resistant to the antibiotic. J. Antimicrob. Chemother. 32, 817–830 (1993).
Fu, C. et al. Biosynthetic studies of telomycin reveal new lipopeptides with enhanced activity. J. Am. Chem. Soc. 137, 7692–7705 (2015).
Tsai, M. et al. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11, 13 (2011).
Machaidze, G., Ziegler, A. & Seelig, J. Specific binding of Ro 09-0198 (cinnamycin) to phosphatidylethanolamine: a thermodynamic analysis. Biochemistry 41, 1965–1971 (2002).
Kawai, F. et al. Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 186, 1475–1483 (2004).
Mileykovskaya, E. & Dowhan, W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091 (2009).
Oliver, P.M. et al. Localization of anionic phospholipids in Escherichia coli cells. J. Bacteriol. 196, 3386–3398 (2014).
Arias-Cartin, R., Grimaldi, S., Arnoux, P., Guigliarelli, B. & Magalon, A. Cardiolipin binding in bacterial respiratory complexes: structural and functional implications. Biochim. Biophys. Acta 1817, 1937–1949 (2012).
Stolpnik, V.G., Solovena, Y.V. & Antonenko, L.I. [Concentration of neothelomycin in the blood of rabbits following intramuscular or oral administration]. Antibiotiki 11, 567–568 (1966).
Needleman, S.B. & Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453 (1970).
Steinbeck, C. et al. The Chemistry Development Kit (CDK): an open-source Java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 43, 493–500 (2003).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Huson, D.H. & Scornavacca, C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 (2012).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Magrane, M. & Consortium, U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, bar009 (2011).
Finn, R.D., Clements, J. & Eddy, S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
This work was funded through a Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery grant (RGPIN 371576-2014) (N.A.M.) and a Joint Programme Initiative on Antimicrobial Resistance funded through the Canadian Institutes of Health Research (CIHR; N.A.M.). C.W.J. is funded through a CIHR Doctoral Research Award. E.D.B and N.A.M. are supported by the Canada Research Chairs Program. The authors acknowledge and thank R. Epand for valuable communications.
Author information
Authors and Affiliations
Contributions
C.W.J. performed telomycin studies, assisted in antibacterial library curation and development of scoring strategies, performed Antibioticome analysis, contributed to study design and wrote the manuscript. M.A.S. developed PRISM, developed the Antibioticome web application, assisted in resistance gene collection and contributed to study design. C.A.D. developed the retrobiosynthetic algorithm, devised scoring strategies for the Antibioticome, developed the Antibioticome web application and contributed to study design. P.N.R. curated the antibacterial library. G.M.C. developed the retrobiosynthetic algorithm and devised scoring strategies for the Antibioticome. C.W. collected resistance genes. S.F. performed microscopy studies. E.D.B. edited the manuscript. J.B. curated the antibacterial library. D.Y.L. assisted in resistance gene collection and curation of the antibacterial library. N.A.M. contributed to study design and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–15 and Supplementary Tables 1–4. (PDF 969 kb)
Supplementary Note
Synthetic Procedures (PDF 4573 kb)
Supplementary Data Set 1
Results from retrobiosynthetic analysis of microbial modular natural product antibacterials (PDF 3440 kb)
Supplementary Data Set 2
Hidden Markov models used by PRISM to detect antibiotic resistance genes (XLSX 66 kb)
Rights and permissions
About this article
Cite this article
Johnston, C., Skinnider, M., Dejong, C. et al. Assembly and clustering of natural antibiotics guides target identification. Nat Chem Biol 12, 233–239 (2016). https://doi.org/10.1038/nchembio.2018
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2018
This article is cited by
-
The antimicrobial effect of a novel peptide LL-1 on Escherichia coli by increasing membrane permeability
BMC Microbiology (2022)
-
Streptomyces coeruleorubidus as a potential biocontrol agent for Newcastle disease virus
BMC Veterinary Research (2022)
-
Outer membrane and phospholipid composition of the target membrane affect the antimicrobial potential of first- and second-generation lipophosphonoxins
Scientific Reports (2021)
-
Mining and unearthing hidden biosynthetic potential
Nature Communications (2021)
-
Towards the sustainable discovery and development of new antibiotics
Nature Reviews Chemistry (2021)