Abstract
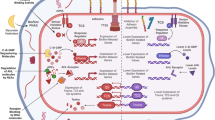
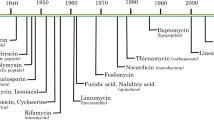
Clinically significant antibiotic resistance has evolved against virtually every antibiotic deployed. Yet the development of new classes of antibiotics has lagged far behind our growing need for such drugs. Rather than focusing on therapeutics that target in vitro viability, much like conventional antibiotics, an alternative approach is to target functions essential for infection, such as virulence factors required to cause host damage and disease. This approach has several potential advantages including expanding the repertoire of bacterial targets, preserving the host endogenous microbiome, and exerting less selective pressure, which may result in decreased resistance. We review new approaches to targeting virulence, discuss their advantages and disadvantages, and propose that in addition to targeting virulence, new antimicrobial development strategies should be expanded to include targeting bacterial gene functions that are essential for in vivo viability. We highlight both new advances in identifying these functions and prospects for antimicrobial discovery targeting this unexploited area.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Palumbi, S.R. Humans as the world's greatest evolutionary force. Science 293, 1786–1790 (2001).
Young, J.A. & Collier, R.J. Anthrax toxin: receptor-binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 76, 243–265 (2007).
Boyden, E.D. & Dietrich, W.F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240–244 (2006).
Duesbery, N.S. et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280, 734–737 (1998).
Vitale, G. et al. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem. Biophys. Res. Commun. 248, 706–711 (1998).
Panchal, R.G. et al. Chemical genetic screening identifies critical pathways in anthrax lethal toxin-induced pathogenesis. Chem. Biol. 14, 245–255 (2007).
During, R.L. et al. Anthrax lethal toxin paralyzes actin-based motility by blocking Hsp27 phosphorylation. EMBO J. 26, 2240–2250 (2007).
Rainey, G.J. & Young, J.A. Antitoxins: novel strategies to target agents of bioterrorism. Nat. Rev. Microbiol. 2, 721–726 (2004).
Shoop, W.L. et al. Anthrax lethal factor inhibition. Proc. Natl. Acad. Sci. USA 102, 7958–7963 (2005).
Turk, B.E. et al. The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Nat. Struct. Mol. Biol. 11, 60–66 (2004).
Krantz, B.A. et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science 309, 777–781 (2005).
Artenstein, A.W. et al. Chloroquine enhances survival in Bacillus anthracis intoxication. J. Infect. Dis. 190, 1655–1660 (2004).
Sanchez, A.M. et al. Amiodarone and bepridil inhibit anthrax toxin entry into host cells. Antimicrob. Agents Chemother 51, 2403–2411 (2007).
Moayeri, M., Wiggins, J.F., Lindeman, R.E. & Leppla, S.H. Cisplatin inhibition of anthrax lethal toxin. Antimicrob. Agents Chemother. 50, 2658–2665 (2006).
Ma, T. et al. Thiazolidinone CFTR inhibitor identified by high-throughput screening blocks cholera toxin-induced intestinal fluid secretion. J. Clin. Invest. 110, 1651–1658 (2002).
King, C.Y. & Barriere, S.L. Analysis of the in vitro interaction between vancomycin and cholestyramine. Antimicrob. Agents Chemother. 19, 326–327 (1981).
Galan, J.E. & Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 (2006).
Kauppi, A.M., Nordfelth, R., Uvell, H., Wolf-Watz, H. & Elofsson, M. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10, 241–249 (2003).
Nordfelth, R., Kauppi, A.M., Norberg, H.A., Wolf-Watz, H. & Elofsson, M. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73, 3104–3114 (2005).
Muschiol, S. et al. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 103, 14566–14571 (2006).
Bailey, L. et al. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581, 587–595 (2007).
Hudson, D.L. et al. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother 51, 2631–2635 (2007).
Stevens, D.L., Gibbons, A.E., Bergstrom, R. & Winn, V. The Eagle effect revisited: efficacy of clindamycin, erythromycin, and penicillin in the treatment of streptococcal myositis. J. Infect. Dis. 158, 23–28 (1988).
Miller, M.B. & Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Parsek, M.R., Val, D.L., Hanzelka, B.L., Cronan, J.E. Jr. & Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96, 4360–4365 (1999).
Dong, Y.H., Xu, J.L., Li, X.Z. & Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97, 3526–3531 (2000).
Dong, Y.H. et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817 (2001).
Yang, F. et al. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 579, 3713–3717 (2005).
Draganov, D.I. et al. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 46, 1239–1247 (2005).
Muh, U. et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50, 3674–3679 (2006).
Geske, G.D., Wezeman, R.J., Siegel, A.P. & Blackwell, H.E. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J. Am. Chem. Soc. 127, 12762–12763 (2005).
Smith, K.M., Bu, Y. & Suga, H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthtic autoinducer analogs. Chem. Biol. 10, 81–89 (2003).
Givskov, M. et al. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178, 6618–6622 (1996).
Manefield, M. et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148, 1119–1127 (2002).
Manefield, M., Welch, M., Givskov, M., Salmond, G.P. & Kjelleberg, S. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 205, 131–138 (2001).
Hentzer, M. et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148, 87–102 (2002).
Hentzer, M. et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815 (2003).
Wu, H. et al. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53, 1054–1061 (2004).
Yates, E.A. et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70, 5635–5646 (2002).
Muh, U. et al. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc. Natl. Acad. Sci. USA 103, 16948–16952 (2006).
Lyon, G.J., Wright, J.S., Christopoulos, A., Novick, R.P. & Muir, T.W. Reversible and specific extracellular antagonism of receptor-histidine kinase signaling. J. Biol. Chem. 277, 6247–6253 (2002).
Mayville, P. et al. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96, 1218–1223 (1999).
Wright, J.S. III, Jin, R. & Novick, R.P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 102, 1691–1696 (2005).
Hung, D.T., Shakhnovich, E.A., Pierson, E. & Mekalanos, J.J. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310, 670–674 (2005).
Gauthier, A. et al. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49, 4101–4109 (2005).
Sauer, F.G. et al. Chaperone-assisted pilus assembly and bacterial attachment. Curr. Opin. Struct. Biol. 10, 548–556 (2000).
Svensson, A. et al. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. ChemBioChem 2, 915–918 (2001).
Pinkner, J.S. et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. USA 103, 17897–17902 (2006).
Lee, Y.M., Almqvist, F. & Hultgren, S.J. Targeting virulence for antimicrobial chemotherapy. Curr. Opin. Pharmacol. 3, 513–519 (2003).
Sassetti, C.M. & Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100, 12989–12994 (2003).
MacRae, C.A. & Peterson, R.T. Zebrafish-based small molecule discovery. Chem. Biol. 10, 901–908 (2003).
Peterson, R.T. et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 22, 595–599 (2004).
Stern, H.M. et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat. Chem. Biol. 1, 366–370 (2005).
Davis, J.M. et al. Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17, 693–702 (2002).
Neely, M.N., Pfeifer, J.D. & Caparon, M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 70, 3904–3914 (2002).
van der Sar, A.M. et al. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 5, 601–611 (2003).
Mukhopadhyay, A. & Peterson, R.T. Fishing for new antimicrobials. Curr. Opin. Chem. Biol. 10, 327–333 (2006).
Moy, T.I. et al. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. USA 103, 10414–10419 (2006).
Payne, D.J., Gwynn, M.N., Holmes, D.J. & Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Schreiber, S.L. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287, 1964–1969 (2000).
Sarac, M.S., Peinado, J.R., Leppla, S.H. & Lindberg, I. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 72, 602–605 (2004).
Acknowledgements
We thank E.J. Rubin, J.E. Gomez and S.A. Stanley for helpful discussions and critical reading of this manuscript and J.S.W. Lee for assistance in preparing figures.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Clatworthy, A., Pierson, E. & Hung, D. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 3, 541–548 (2007). https://doi.org/10.1038/nchembio.2007.24
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2007.24
This article is cited by
-
Antibacterial Activity of Silver Nanoparticles Derived from Extracellular Extract of Enterococcus aerogenes Against Dental Disease Bacteria Isolated
Regenerative Engineering and Translational Medicine (2024)
-
Accurate and rapid antibiotic susceptibility testing using a machine learning-assisted nanomotion technology platform
Nature Communications (2024)
-
Development of a smart pH-responsive nano-polymer drug, 2-methoxy-4-vinylphenol conjugate against the intestinal pathogen, Vibrio cholerae
Scientific Reports (2023)
-
Efficacy of ceftiofur N-acyl homoserine lactonase niosome in the treatment of multi-resistant Klebsiella pneumoniae in broilers
Veterinary Research Communications (2023)
-
Deep Learning Framework for Predicting Essential Proteins with Temporal Convolutional Networks
Journal of Shanghai Jiaotong University (Science) (2023)