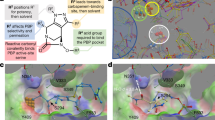
Abstract
β-lactam antibiotics, including penicillins and cephalosporins, inhibit penicillin-binding proteins (PBPs), which are essential for bacterial cell wall biogenesis. Pathogenic bacteria have evolved efficient antibiotic resistance mechanisms that, in Gram-positive bacteria, include mutations to PBPs that enable them to avoid β-lactam inhibition1. Lactivicin (LTV; 1) contains separate cycloserine and γ-lactone rings and is the only known natural PBP inhibitor that does not contain a β-lactam2,3,4. Here we show that LTV and a more potent analog, phenoxyacetyl-LTV (PLTV; 2), are active against clinically isolated, penicillin-resistant Streptococcus pneumoniae strains. Crystallographic analyses of S. pneumoniae PBP1b reveal that LTV and PLTV inhibition involves opening of both monocyclic cycloserine and γ-lactone rings. In PBP1b complexes, the ring-derived atoms from LTV and PLTV show a notable structural convergence with those derived from a complexed cephalosporin (cefotaxime; 3). The structures imply that derivatives of LTV will be useful in the search for new antibiotics with activity against β-lactam–resistant bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Macheboeuf, P., Contreras-Martel, C., Job, V., Dideberg, O. & Dessen, A. Penicillin Binding Proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30, 673–691 (2006).
Nozaki, Y. et al. Binding of a non-β-lactam antibiotic to penicillin-binding proteins. Nature 325, 179–180 (1987).
Nozaki, Y., Katayama, N., Harada, S., Ono, H. & Okazaki, H. Lactivicin, a naturally occuring non-β-lactam antibiotic having β-lactam-like action: Biological activities and mode of action. J. Antibiot. (Tokyo) 42, 84–93 (1989).
Harada, S. et al. Structure of lactivicin, an antibiotic having a new nucleus and similar biological activities to β-lactam antibiotics. Tetrahedr. Lett. 27, 6229–6232 (1986).
Schuchat, A. et al. Bacterial meningitis in the United States in 1995. Active Surveillance Team. N. Engl. J. Med. 337, 970–976 (1997).
Bronzwaer, S.L. et al. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg. Infect. Dis. 8, 278–282 (2002).
Page, M.I. The Chemistry of β-Lactams (ed. Page, M.I.) (Blackie, Glasgow, 1992).
Harada, S. et al. Chemistry of a new antibiotic: Lactivicin. Tetrahedron 44, 6589–6606 (1988).
Inui, T., Oshida, T., Endo, T. & Matsushita, T. Potent bacteriolytic activity of ritipenem associated with a characteristic profile of affinities for penicillin-binding proteins of Haemophilus influenzae. Antimicrob. Agents Chemother. 43, 2534–2537 (1999).
Hakenbeck, R. et al. Penicillin-binding proteins in beta-lactam-resistant Streptococcus pneumoniae. Microb. Drug Resist. 5, 91–99 (1999).
Chesnel, L. et al. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to β-lactams of resistant strains. J. Biol. Chem. 278, 44448–44456 (2003).
Macheboeuf, P. et al. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 102, 577–582 (2005).
Imming, P., Klar, B. & Dix, D. Hydrolytic stability versus ring size in lactams: Implications for the development of lactam antibiotics and other serine protease inhibitors. J. Med. Chem. 43, 4328–4331 (2000).
Wilmouth, R.C. et al. Mechanistic insights into the inhibition of serine proteases by monocyclic lactams. Biochemistry 38, 7989–7998 (1999).
Brown, R.P.A., Aplin, R.T. & Schofield, C.J. Inhibition of TEM-2 β-lactamase from Escherichia coli by clavulanic acid: Observation of intermediates by electrospray ionization mass spectrometry. Biochemistry 35, 12421–12432 (1996).
Imtiaz, U. et al. Inactivation of class-A β-lactamases by clavulanic acid—the role of arginine-244 in a proposed nonconcerted sequence of events. J. Am. Chem. Soc. 115, 4435–4442 (1993).
Llinás, A. et al. Inactivation of bacterial DD-peptidase by β-sultams. Biochemistry 44, 7738–7746 (2005).
Lakaye, B. et al. Synthesis, purification and kinetic properties of fluorescein-labelled penicillins. Biochem. J. 300, 141–145 (1994).
Carapito, R., Chesnel, L., Vernet, T. & Zapun, A. Pneumococcal β-lactam resistance due to a conformational change in penicillin-binding protein 2x. J. Biol. Chem. 281, 1771–1777 (2006).
Natsugari, H., Kawano, Y., Morimoto, A. & Yoshioka, K. Antibiotic 2-(3-oxo-2-isoxazolidinyl)-5-oxo-2-tetrahydrofuran-carboxylates. US Patent 4,851,422 (1989).
Ghuysen, J.M., Frère, J.M., Leyh-Bouille, M., Nguyen-Disteche, M. & Coyette, J. Active-site-serine D-alanyl-D-alanine-cleaving-peptidase-catalysed acyl-transfer reactions. Procedures for studying the penicillin-binding proteins of bacterial plasma membranes. Biochem. J. 235, 159–165 (1986).
Adam, M., Damblon, C., Plaitin, B., Christiaens, L. & Frère, J.M. Chromogenic depsipeptide substrates for beta-lactamases and penicillin-sensitive DD-peptidases. Biochem. J. 270, 525–529 (1990).
De Meester, F. et al. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem. Pharmacol. 36, 2393–2403 (1987).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. Sect. D Biol. Crystallogr. 57, 1367–1372 (2001).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004).
Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 760–763 (1994).
Brünger, A. Crystallography & NMR system: A new software for macromolecular structure determination. Acta Crystallogr. Sect. D. Biol. Crystallogr. 54, 905–921 (1998).
Acknowledgements
We thank the ESRF ID14 and ID23 beamline staff for help with data collection, D. Lemaire (IBS) for mass spectrometric assays, J. Croizé (Hopital Universitaire de Grenoble) for drug-resistant pneumococcal strains, R. Carapito for the purification of PBP2x (5204) and O. Dideberg for support. We thank J.M. Frère (ULG) for useful discussions. The work was funded by the European Commission LSHM-CT-2004-512138 (EUR-INTAFAR).
Author information
Authors and Affiliations
Contributions
P.M. and A.D. carried out the crystallographic analyses; D.S.F. and T.B. Jr. carried out the synthetic studies; A.Z., A.L., P.M. and B.J. carried out microbiological and kinetic analyses; A.D., D.S.F. and C.J.S. designed the study, analyzed data and, together with the other authors, wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figure1, Supplementary Table1 and Supplementary Methods (PDF 261 kb)
Rights and permissions
About this article
Cite this article
Macheboeuf, P., Fischer, D., Brown, T. et al. Structural and mechanistic basis of penicillin-binding protein inhibition by lactivicins. Nat Chem Biol 3, 565–569 (2007). https://doi.org/10.1038/nchembio.2007.21
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2007.21